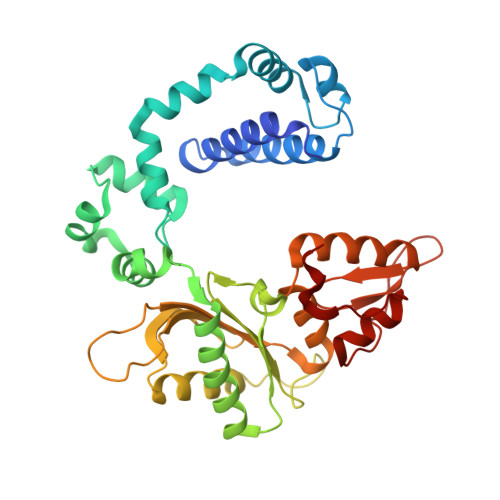
Unfavorable Electrostatic and Steric Interactions in DNA Polymerase beta E295K Mutant Interfere with the Enzyme s Pathway
Li, Y., Gridley, C.L., Jaeger, J., Sweasy, J.B., Schlick, T.(2012) J Am Chem Soc 134: 9999-10010
- PubMed: 22651551
- DOI: https://doi.org/10.1021/ja300361r
- Primary Citation of Related Structures:
3V72 - PubMed Abstract:
Mutations in DNA polymerase β (pol β) have been associated with approximately 30% of human tumors. The E295K mutation of pol β has been linked to gastric carcinoma via interference with base excision repair. To interpret the different behavior of E295K as compared to wild-type pol β in atomic and energetic detail, we resolve a binary crystal complex of E295K at 2.5 Å and apply transition path sampling (TPS) to delineate the closing pathway of the E295K pol β mutant. Conformational changes are important components in the enzymatic pathway that lead to and ready the enzyme for the chemical reaction. Our analyses show that the closing pathway of E295K mutant differs from the wild-type pol β in terms of the individual transition states along the pathway, associated energies, and the active site conformation in the final closed form of the mutant. In particular, the closed state of E295K has a more distorted active site than the active site in the wild-type pol β. In addition, the total energy barrier in the conformational closing pathway is 65 ± 11 kJ/mol, much higher than that estimated for both correct (e.g., G:C) and incorrect (e.g., G:A) wild-type pol β systems (42 ± 8 and 45 ± 7 kJ/mol, respectively). In particular, the rotation of Arg258 is the rate-limiting step in the conformational pathway of E295K due to unfavorable electrostatic and steric interactions. The distorted active site in the closed relative to open state and the high energy barrier in the conformational pathway may explain in part why the E295K mutant is observed to be inactive. Interestingly, however, following the closing of the thumb but prior to the rotation of Arg258, the E295K mutant complex has a similar energy level as compared to the wild-type pol β. This suggests that the E295K mutant may associate with DNA with similar affinity, but it may be hampered in continuing the process of chemistry. Supporting experimental data come from the observation that the catalytic activity of wild-type pol β is hampered when E295K is present: this may arise from the competition between E295K and wild-type enzyme for the DNA. These combined results suggest that the low insertion efficiency of E295K mutant as compared to wild-type pol β may be related to a closed form distorted by unfavorable electrostatic and steric interactions between Arg258 and other key residues. The active site is thus less competent for proceeding to the chemical reaction, which may also involve a higher reaction barrier than the wild-type or may not be possible in this mutant. Our analysis also suggests further experiments for other mutants to test the above hypothesis and dissect the roles of steric and electrostatic factors on enzyme behavior.
- Department of Chemistry and Courant Institute of Mathematical Sciences, New York University, 251 Mercer Street, New York, New York 10012, USA.
Organizational Affiliation: