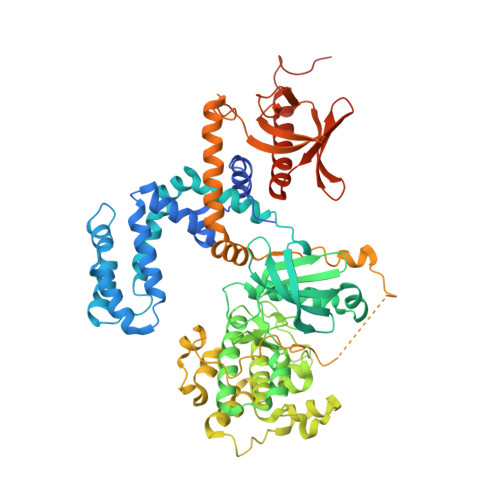
Paroxetine is a direct inhibitor of g protein-coupled receptor kinase 2 and increases myocardial contractility.
Thal, D.M., Homan, K.T., Chen, J., Wu, E.K., Hinkle, P.M., Huang, Z.M., Chuprun, J.K., Song, J., Gao, E., Cheung, J.Y., Sklar, L.A., Koch, W.J., Tesmer, J.J.(2012) ACS Chem Biol 7: 1830-1839
- PubMed: 22882301
- DOI: https://doi.org/10.1021/cb3003013
- Primary Citation of Related Structures:
3V5W - PubMed Abstract:
G protein-coupled receptor kinase 2 (GRK2) is a well-established therapeutic target for the treatment of heart failure. Herein we identify the selective serotonin reuptake inhibitor (SSRI) paroxetine as a selective inhibitor of GRK2 activity both in vitro and in living cells. In the crystal structure of the GRK2·paroxetine-Gβγ complex, paroxetine binds in the active site of GRK2 and stabilizes the kinase domain in a novel conformation in which a unique regulatory loop forms part of the ligand binding site. Isolated cardiomyocytes show increased isoproterenol-induced shortening and contraction amplitude in the presence of paroxetine, and pretreatment of mice with paroxetine before isoproterenol significantly increases left ventricular inotropic reserve in vivo with no significant effect on heart rate. Neither is observed in the presence of the SSRI fluoxetine. Our structural and functional results validate a widely available drug as a selective chemical probe for GRK2 and represent a starting point for the rational design of more potent and specific GRK2 inhibitors.
- Life Sciences Institute and the Department of Pharmacology, University of Michigan, Ann Arbor, Michigan 48109, United States.
Organizational Affiliation: