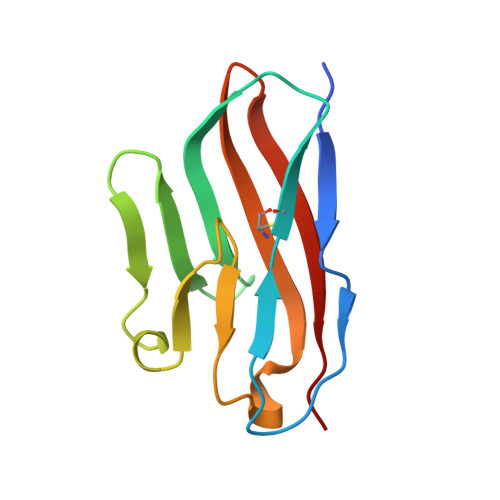
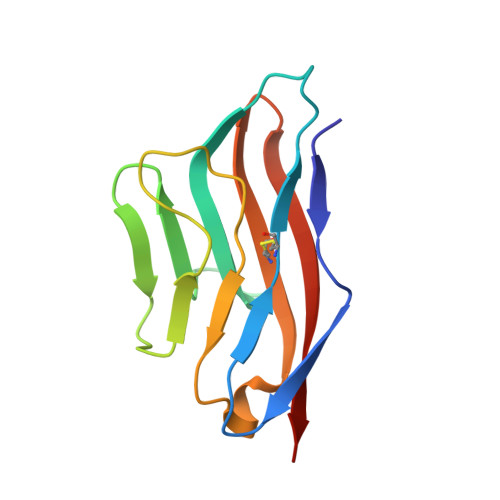
Structure of TIGIT immunoreceptor bound to poliovirus receptor reveals a cell-cell adhesion and signaling mechanism that requires cis-trans receptor clustering.
Stengel, K.F., Harden-Bowles, K., Yu, X., Rouge, L., Yin, J., Comps-Agrar, L., Wiesmann, C., Bazan, J.F., Eaton, D.L., Grogan, J.L.(2012) Proc Natl Acad Sci U S A 109: 5399-5404
- PubMed: 22421438
- DOI: https://doi.org/10.1073/pnas.1120606109
- Primary Citation of Related Structures:
3UCR, 3UDW - PubMed Abstract:
Nectins (nectin1-4) and Necls [nectin-like (Necl1-5)] are Ig superfamily cell adhesion molecules that regulate cell differentiation and tissue morphogenesis. Adherens junction formation and subsequent cell-cell signaling is initiated by the assembly of higher-order receptor clusters of cognate molecules on juxtaposed cells. However, the structural and mechanistic details of signaling cluster formation remain unclear. Here, we report the crystal structure of poliovirus receptor (PVR)/Nectin-like-5/CD155) in complex with its cognate immunoreceptor ligand T-cell-Ig-and-ITIM-domain (TIGIT). The TIGIT/PVR interface reveals a conserved specific "lock-and-key" interaction. Notably, two TIGIT/PVR dimers assemble into a heterotetramer with a core TIGIT/TIGIT cis-homodimer, each TIGIT molecule binding one PVR molecule. Structure-guided mutations that disrupt the TIGIT/TIGIT interface limit both TIGIT/PVR-mediated cell adhesion and TIGIT-induced PVR phosphorylation in primary dendritic cells. Our data suggest a cis-trans receptor clustering mechanism for cell adhesion and signaling by the TIGIT/PVR complex and provide structural insights into how the PVR family of immunoregulators function.
- Department of Structural Biology, Genentech, Inc., South San Francisco, CA 94080, USA.
Organizational Affiliation: