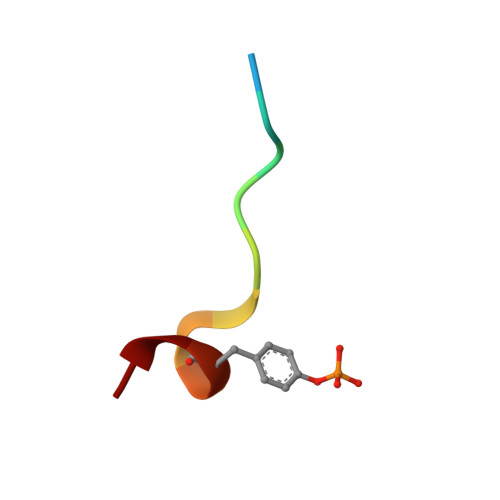
Molecular design and structure-activity relationships leading to the potent, selective, and orally active thrombin active site inhibitor BMS-189664.
Das, J., Kimball, S.D., Hall, S.E., Han, W.C., Iwanowicz, E., Lin, J., Moquin, R.V., Reid, J.A., Sack, J.S., Malley, M.F., Chang, C.Y., Chong, S., Wang-Iverson, D.B., Roberts, D.G., Seiler, S.M., Schumacher, W.A., Ogletree, M.L.(2002) Bioorg Med Chem Lett 12: 45-49
- PubMed: 11738570
- DOI: https://doi.org/10.1016/s0960-894x(01)00667-9
- Primary Citation of Related Structures:
3TU7 - PubMed Abstract:
A series of structurally novel small molecule inhibitors of human alpha-thrombin was prepared to elucidate their structure-activity relationships (SARs), selectivity and activity in vivo. BMS-189664 (3) is identified as a potent, selective, and orally active reversible inhibitor of human alpha-thrombin which is efficacious in vivo in a mouse lethality model, and at inhibiting both arterial and venous thrombosis in cynomolgus monkey models.
- Bristol-Myers Squibb Pharmaceutical Research Institute, Princeton, NJ 08543-4000, USA. jagabandhu.das@bms.com
Organizational Affiliation: