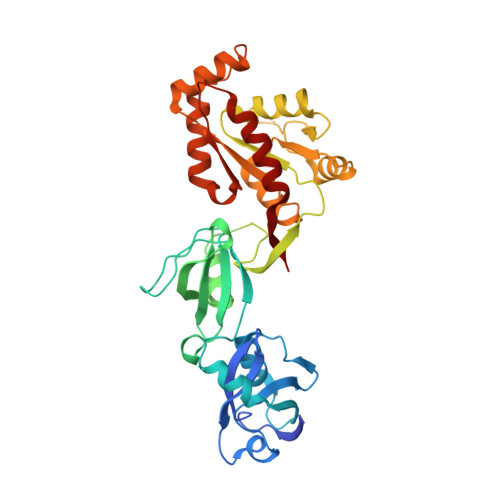
The Src Homology 3 Domain Is Required for Junctional Adhesion Molecule Binding to the Third PDZ Domain of the Scaffolding Protein ZO-1.
Nomme, J., Fanning, A.S., Caffrey, M., Lye, M.F., Anderson, J.M., Lavie, A.(2011) J Biological Chem 286: 43352-43360
- PubMed: 22030391
- DOI: https://doi.org/10.1074/jbc.M111.304089
- Primary Citation of Related Structures:
3TSV, 3TSW, 3TSZ - PubMed Abstract:
Tight junctions are cell-cell contacts that regulate the paracellular flux of solutes and prevent pathogen entry across cell layers. The assembly and permeability of this barrier are dependent on the zonula occludens (ZO) membrane-associated guanylate kinase (MAGUK) proteins ZO-1, -2, and -3. MAGUK proteins are characterized by a core motif of protein-binding domains that include a PDZ domain, a Src homology 3 (SH3) domain, and a region of homology to guanylate kinase (GUK); the structure of this core motif has never been determined for any MAGUK. To better understand how ZO proteins organize the assembly of protein complexes we have crystallized the entire PDZ3-SH3-GUK core motif of ZO-1. We have also crystallized this core motif in complex with the cytoplasmic tail of the ZO-1 PDZ3 ligand, junctional adhesion molecule A (JAM-A) to determine how the activity of different domains is coordinated. Our study shows a new feature for PDZ class II ligand binding that implicates the two highly conserved Phe(-2) and Ser(-3) residues of JAM. Our x-ray structures and NMR experiments also show for the first time a role for adjacent domains in the binding of ligands to PDZ domains in the MAGUK proteins family.
- Department of Biochemistry and Molecular Genetics, University of Illinois, Chicago, Illinois 60607, USA.
Organizational Affiliation: