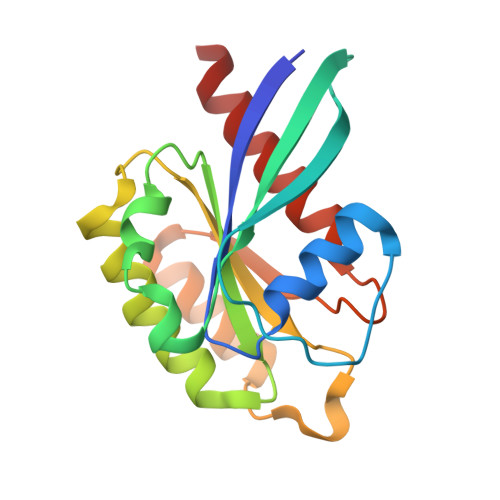
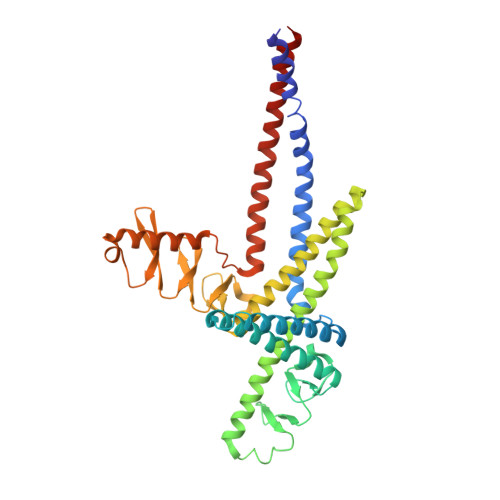
Protein LidA from Legionella is a Rab GTPase supereffector.
Schoebel, S., Cichy, A.L., Goody, R.S., Itzen, A.(2011) Proc Natl Acad Sci U S A 108: 17945-17950
- PubMed: 22011575
- DOI: https://doi.org/10.1073/pnas.1113133108
- Primary Citation of Related Structures:
3TNF - PubMed Abstract:
The causative agent of Legionnaires disease, Legionella pneumophila, injects several hundred proteins into the cell it infects, many of which interfere with or exploit vesicular transport processes. One of these proteins, LidA, has been described as a Rab effector (i.e., a molecule that interacts preferentially with the GTP-bound form of Rab). We describe here the structure and biochemistry of a complex between the Rab-binding domain of LidA and active Rab8a. LidA displays structural peculiarities in binding to Rab8a, forming a considerably extended interface in comparison to known mammalian Rab effectors, and involving regions of the GTPase that are not seen in other Rab:effector complexes. In keeping with this extended binding interface, which involves four α-helices and two pillar-like structures of LidA, the stability of LidA-Rab interactions is dramatically greater than for other such complexes. For Rab1b and Rab8a, these affinities are extraordinarily high, but for the more weakly bound Rab6a, K(d) values of 4 nM for the inactive and 30 pM for the active form were found. Rab1b and Rab8a appear to bind LidA with K(d) values in the low picomolar range, making LidA a Rab supereffector.
- Department of Physical Biochemistry, Max-Planck-Institute of Molecular Physiology, Otto-Hahn-Strasse 11, Dortmund 44227, Germany.
Organizational Affiliation: