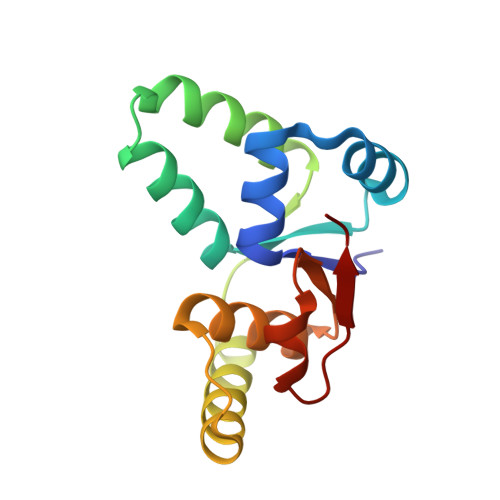
Crystal Structure of the VapBC Toxin-Antitoxin Complex from Shigella flexneri Reveals a Hetero-Octameric DNA-Binding Assembly.
Dienemann, C., Boggild, A., Winther, K.S., Gerdes, K., Brodersen, D.E.(2011) J Mol Biology 414: 713-722
- PubMed: 22037005
- DOI: https://doi.org/10.1016/j.jmb.2011.10.024
- Primary Citation of Related Structures:
3TND - PubMed Abstract:
Toxin-antitoxin (TA) loci are common in archaea and prokaryotes and allow cells to rapidly adapt to changing environmental conditions through release of active regulators of metabolism. Many toxins are endonucleases that target cellular mRNA and tRNAs, while the antitoxins tightly wrap around the toxins to inhibit them under normal circumstances. The antitoxins also bind to operators in the promoter regions of the cognate TA operon and thereby regulate transcription. For enteric vapBC TA loci, the VapC toxins specifically cleave tRNA(fMet) and thus down-regulate protein synthesis. Here, we describe the crystal structure of the intact Shigella flexneri VapBC TA complex, determined to 2.7 Å resolution. Both in solution and in the crystal structure, four molecules of each protein combine to form a large and globular hetero-octameric assembly with SpoVT/AbrB-type DNA-binding domains at each end and a total molecular mass of about 100 kDa. The structure gives new insights into the inhibition of VapC toxins by VapB and provides the molecular basis for understanding transcriptional regulation through VapB dimerization.
- Centre for mRNP Biogenesis and Metabolism, Department of Molecular Biology and Genetics, Aarhus University, Gustav Wieds Vej 10C, DK-8000 Aarhus C, Denmark.
Organizational Affiliation: