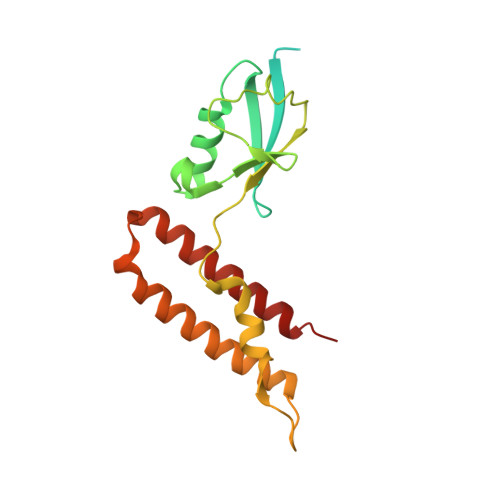
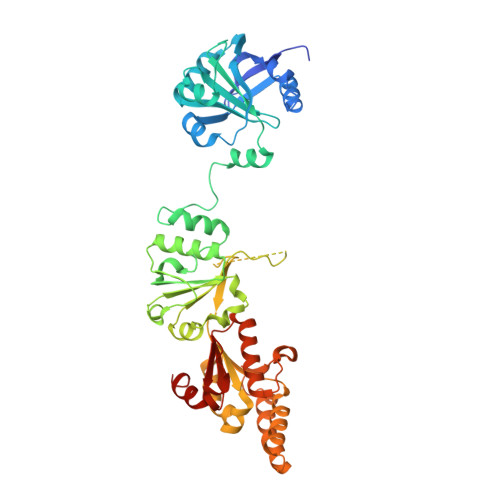
The Chp1-Tas3 core is a multifunctional platform critical for gene silencing by RITS.
Schalch, T., Job, G., Shanker, S., Partridge, J.F., Joshua-Tor, L.(2011) Nat Struct Mol Biol 18: 1351-1357
- PubMed: 22081013
- DOI: https://doi.org/10.1038/nsmb.2151
- Primary Citation of Related Structures:
3TIX - PubMed Abstract:
RNA interference (RNAi) is critical for the assembly of heterochromatin at Schizosaccharomyces pombe centromeres. Central to this process is the RNA-induced initiation of transcriptional gene silencing (RITS) complex, which physically anchors small noncoding RNAs to chromatin. RITS includes Ago1, the chromodomain protein Chp1, and Tas3, which forms a bridge between Chp1 and Ago1. Chp1 is a large protein with no recognizable domains, apart from its chromodomain. Here we describe how the structured C-terminal half of Chp1 binds the Tas3 N-terminal domain, revealing the tight association of Chp1 and Tas3. The structure also shows a PIN domain at the C-terminal tip of Chp1 that controls subtelomeric transcripts through a post-transcriptional mechanism. We suggest that the Chp1-Tas3 complex provides a solid and versatile platform to recruit both RNAi-dependent and RNAi-independent gene-silencing pathways for locus-specific regulation of heterochromatin.
- Keck Structural Biology Laboratory, Cold Spring Harbor Laboratory, Cold Spring Harbor, New York, USA.
Organizational Affiliation: