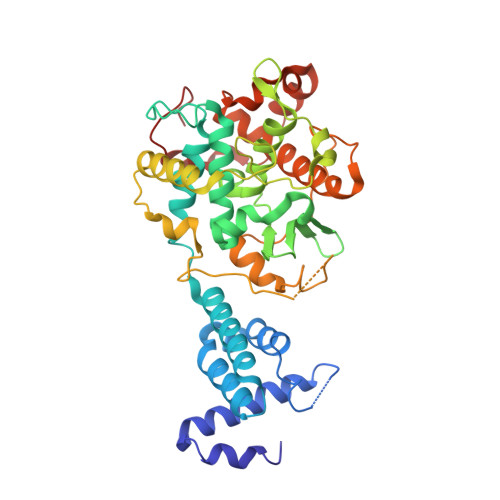
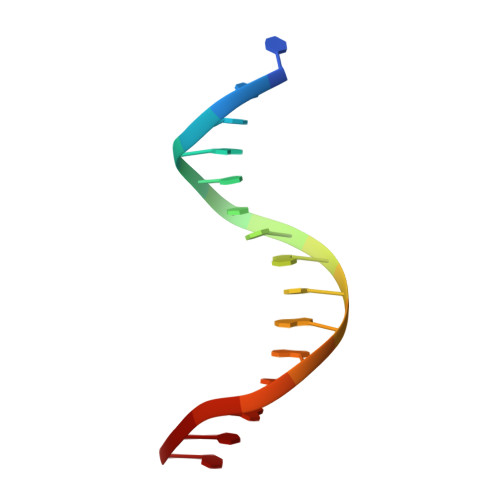
Ndc10 is a platform for inner kinetochore assembly in budding yeast.
Cho, U.S., Harrison, S.C.(2011) Nat Struct Mol Biol 19: 48-55
- PubMed: 22139014
- DOI: https://doi.org/10.1038/nsmb.2178
- Primary Citation of Related Structures:
3SQI, 3T79 - PubMed Abstract:
Kinetochores link centromeric DNA to spindle microtubules and ensure faithful chromosome segregation during mitosis. In point-centromere yeasts, the CBF3 complex Skp1-Ctf13-(Cep3)(2)-(Ndc10)(2) recognizes a conserved centromeric DNA element through contacts made by Cep3 and Ndc10. We describe here the five-domain organization of Kluyveromyces lactis Ndc10 and the structure at 2.8 Å resolution of domains I-II (residues 1-402) bound to DNA. The structure resembles tyrosine DNA recombinases, although it lacks both endonuclease and ligase activities. Structural and biochemical data demonstrate that each subunit of the Ndc10 dimer binds a separate fragment of DNA, suggesting that Ndc10 stabilizes a DNA loop at the centromere. We describe in vitro association experiments showing that specific domains of Ndc10 interact with each of the known inner-kinetochore proteins or protein complexes in budding yeast. We propose that Ndc10 provides a central platform for inner-kinetochore assembly.
- Jack and Eileen Connors Structural Biology Laboratory and Howard Hughes Medical Institute, Department of Biological Chemistry and Molecular Pharmacology, Harvard Medical School, Boston, Massachusetts, USA.
Organizational Affiliation: