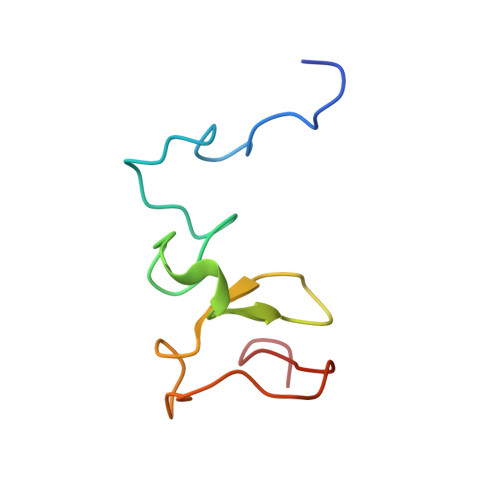
UHRF1 double tudor domain and the adjacent PHD finger act together to recognize K9me3-containing histone H3 tail
Xie, S., Jakoncic, J., Qian, C.M.(2012) J Mol Biology 415: 318-328
- PubMed: 22100450
- DOI: https://doi.org/10.1016/j.jmb.2011.11.012
- Primary Citation of Related Structures:
3T6R - PubMed Abstract:
Human multi-domain-containing protein UHRF1 has recently been extensively characterized as a key epigenetic regulator for maintaining DNA methylation patterns. UHRF1 SRA domain preferentially binds to hemimethylated CpG sites, and double Tudor domain has been implicated in recognizing H3K9me3 mark, but the role of the adjacent PHD finger remains unclear. Here, we report the high-resolution crystal structure of UHRF1 PHD finger in complex with N-terminal tail of histone H3. We found that the preceding zinc-Cys4 knuckle is indispensable for the PHD finger of UHRF1 to recognize the first four unmodified residues of histone H3 N-terminal tail. Quantitative binding studies indicated that UHRF1 PHD finger (including the preceding zinc-Cys4 knuckle) acts together with the adjacent double Tudor domain to specifically recognize the H3K9me3 mark. Combinatorial recognition of H3K9me3-containing histone H3 tail by UHRF1 PHD finger and double Tudor domain may play a role in establishing and maintaining histone H3K9 methylation patterns during the cell cycle.
- Department of Biochemistry, The University of Hong Kong, Hong Kong, China.
Organizational Affiliation: