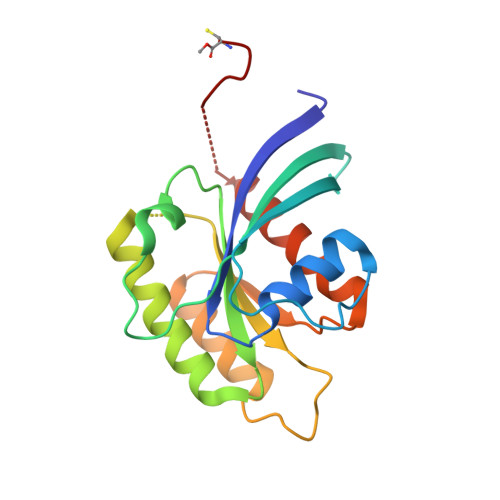
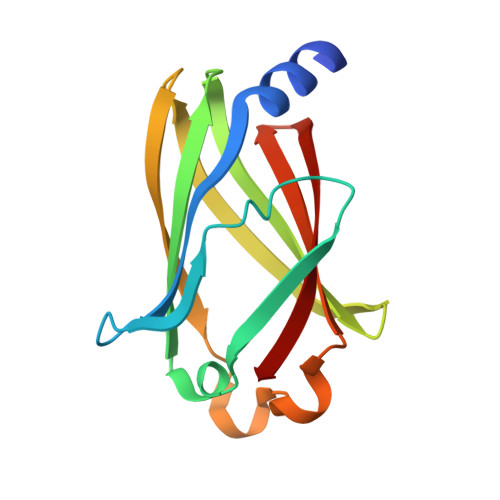
Arl2-GTP and Arl3-GTP regulate a GDI-like transport system for farnesylated cargo.
Ismail, S.A., Chen, Y.X., Rusinova, A., Chandra, A., Bierbaum, M., Gremer, L., Triola, G., Waldmann, H., Bastiaens, P.I., Wittinghofer, A.(2011) Nat Chem Biol 7: 942-949
- PubMed: 22002721
- DOI: https://doi.org/10.1038/nchembio.686
- Primary Citation of Related Structures:
3T5G, 3T5I - PubMed Abstract:
Lipidated Rho and Rab GTP-binding proteins are transported between membranes in complex with solubilizing factors called 'guanine nucleotide dissociation inhibitors' (GDIs). Unloading from GDIs using GDI displacement factors (GDFs) has been proposed but remains mechanistically elusive. PDEδ is a putative solubilizing factor for several prenylated Ras-subfamily proteins. Here we report the structure of fully modified farnesylated Rheb-GDP in complex with PDEδ. The structure explains the nucleotide-independent binding of Rheb to PDEδ and the relaxed specificity of PDEδ. We demonstrate that the G proteins Arl2 and Arl3 act in a GTP-dependent manner as allosteric release factors for farnesylated cargo. We thus describe a new transport system for farnesylated G proteins involving a GDI-like molecule and an unequivocal GDF. Considering the importance of PDEδ for proper Ras and Rheb signaling, this study is instrumental in developing a new target for anticancer therapy.
- Structural Biology Group, Max Planck Institute for Molecular Physiology, Dortmund, Germany.
Organizational Affiliation: