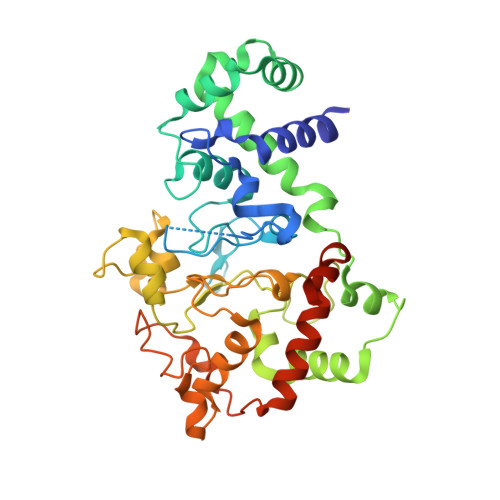
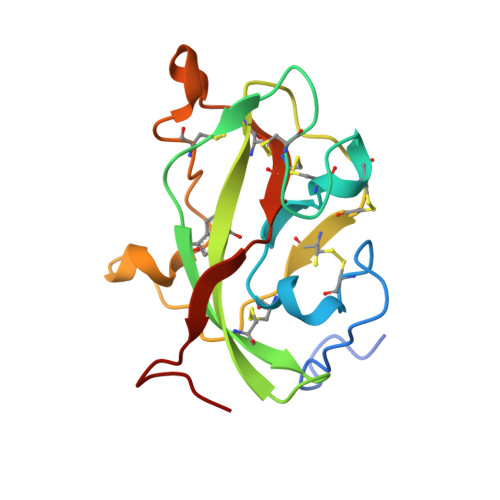
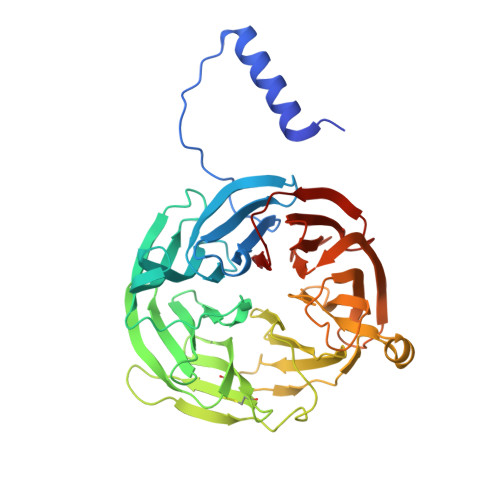
Proline 107 is a major determinant in maintaining the structure of the distal pocket and reactivity of the high-spin heme of MauG.
Feng, M., Jensen, L.M., Yukl, E.T., Wei, X., Liu, A., Wilmot, C.M., Davidson, V.L.(2012) Biochemistry 51: 1598-1606
- PubMed: 22299652
- DOI: https://doi.org/10.1021/bi201882e
- Primary Citation of Related Structures:
3SJL, 3SLE, 3SVW - PubMed Abstract:
The diheme enzyme MauG catalyzes a six-electron oxidation required for posttranslational modification of a precursor of methylamine dehydrogenase (preMADH) to complete the biosynthesis of its protein-derived tryptophan tryptophylquinone (TTQ) cofactor. Crystallographic studies had shown that Pro107, which resides in the distal pocket of the high-spin heme of MauG, changes conformation upon binding of CO or NO to the heme iron. In this study, Pro107 was converted to Cys, Val, and Ser by site-directed mutagenesis. The structures of each of these MauG mutant proteins in complex with preMADH were determined, as were their physical and catalytic properties. P107C MauG was inactive, and the crystal structure revealed that Cys107 had been oxidatively modified to a sulfinic acid. Mass spectrometry revealed that this modification was present prior to crystallization. P107V MauG exhibited spectroscopic and catalytic properties that were similar to those of wild-type MauG, but P107V MauG was more susceptible to oxidative damage. The P107S mutation caused a structural change that resulted in the five-coordinate high-spin heme being converted to a six-coordinate heme with a distal axial ligand provided by Glu113. EPR and resonance Raman spectroscopy revealed this heme remained high-spin but with greatly increased rhombicity as compared to that of the axial signal of wild-type MauG. P107S MauG was resistant to reduction by dithionite and reaction with H(2)O(2) and unable to catalyze TTQ biosynthesis. These results show that the presence of Pro107 is critical in maintaining the proper structure of the distal heme pocket of the high-spin heme of MauG, allowing exogenous ligands to bind and directing the reactivity of the heme-activated oxygen during catalysis, thus minimizing the oxidation of other residues of MauG.
- Department of Chemistry, Tougaloo College, Tougaloo, Mississippi 39174, United States.
Organizational Affiliation: