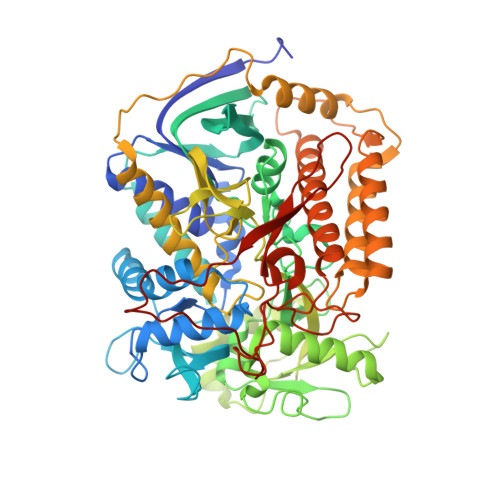
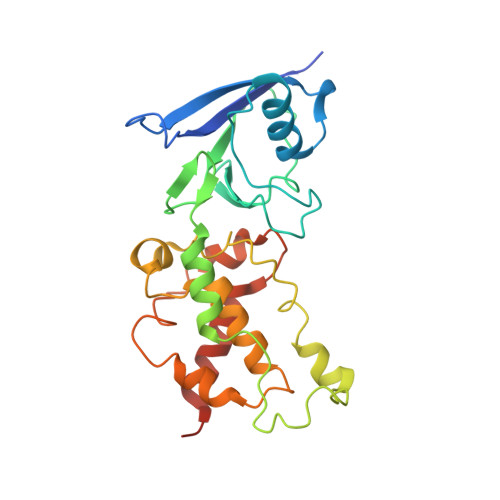
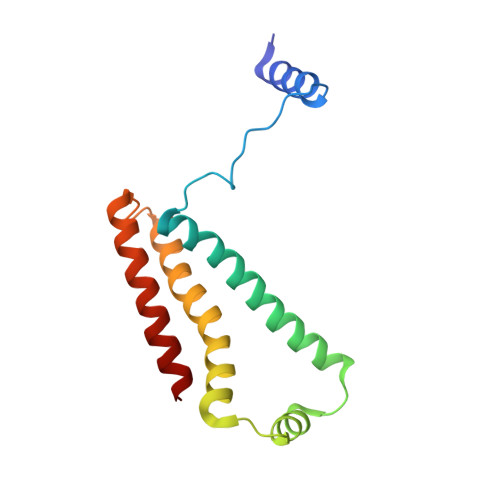
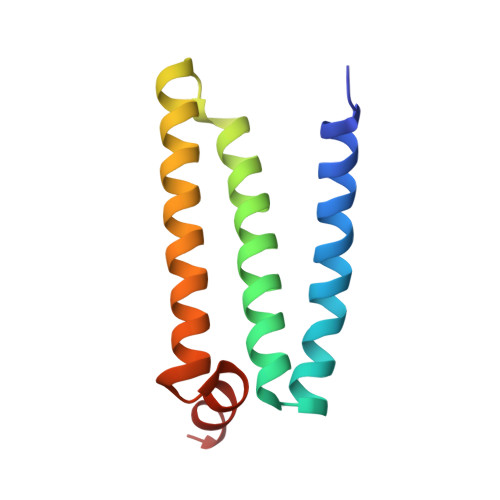
Thiabendazole inhibits ubiquinone reduction activity of mitochondrial respiratory complex II via a water molecule mediated binding feature.
Zhou, Q., Zhai, Y., Lou, J., Liu, M., Pang, X., Sun, F.(2011) Protein Cell 2: 531-542
- PubMed: 21822798
- DOI: https://doi.org/10.1007/s13238-011-1079-1
- Primary Citation of Related Structures:
3SFD, 3SFE - PubMed Abstract:
The mitochondrial respiratory complex II or succinate: ubiquinone oxidoreductase (SQR) is a key membrane complex in both the tricarboxylic acid cycle and aerobic respiration. Five disinfectant compounds were investigated with their potent inhibition effects on the ubiquinone reduction activity of the porcine mitochondrial SQR by enzymatic assay and crystallography. Crystal structure of the SQR bound with thiabendazole (TBZ) reveals a different inhibitor-binding feature at the ubiquinone binding site where a water molecule plays an important role. The obvious inhibitory effect of TBZ based on the biochemical data (IC(50) ~100 μmol/L) and the significant structure-based binding affinity calculation (~94 μmol/L) draw the suspicion of using TBZ as a good disinfectant compound for nematode infections treatment and fruit storage.
- National Laboratory of Biomacromolecules, Institute of Biophysics, Chinese Academy of Sciences, Beijing 100101, China.
Organizational Affiliation: