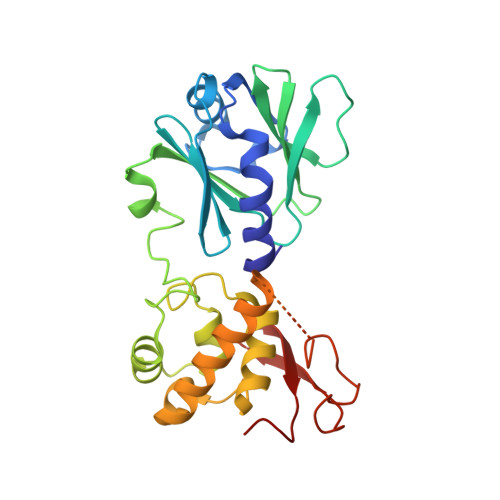
Strandwise translocation of a DNA glycosylase on undamaged DNA.
Qi, Y., Nam, K., Spong, M.C., Banerjee, A., Sung, R.J., Zhang, M., Karplus, M., Verdine, G.L.(2012) Proc Natl Acad Sci U S A 109: 1086-1091
- PubMed: 22219368
- DOI: https://doi.org/10.1073/pnas.1111237108
- Primary Citation of Related Structures:
3SAR, 3SAS, 3SAT, 3SAU, 3SAV, 3SAW, 3SBJ - PubMed Abstract:
Base excision repair of genotoxic nucleobase lesions in the genome is critically dependent upon the ability of DNA glycosylases to locate rare sites of damage embedded in a vast excess of undamaged DNA, using only thermal energy to fuel the search process. Considerable interest surrounds the question of how DNA glycosylases translocate efficiently along DNA while maintaining their vigilance for target damaged sites. Here, we report the observation of strandwise translocation of 8-oxoguanine DNA glycosylase, MutM, along undamaged DNA. In these complexes, the protein is observed to translocate by one nucleotide on one strand while remaining untranslocated on the complementary strand. We further report that alterations of single base-pairs or a single amino acid substitution (R112A) can induce strandwise translocation. Molecular dynamics simulations confirm that MutM can translocate along DNA in a strandwise fashion. These observations reveal a previously unobserved mode of movement for a DNA-binding protein along the surface of DNA.
- Program in Biophysics, Harvard Medical School, 240 Longwood Avenue, Boston, MA 02115, USA.
Organizational Affiliation: