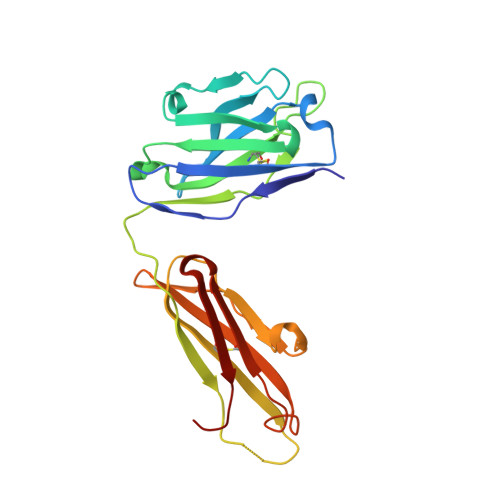
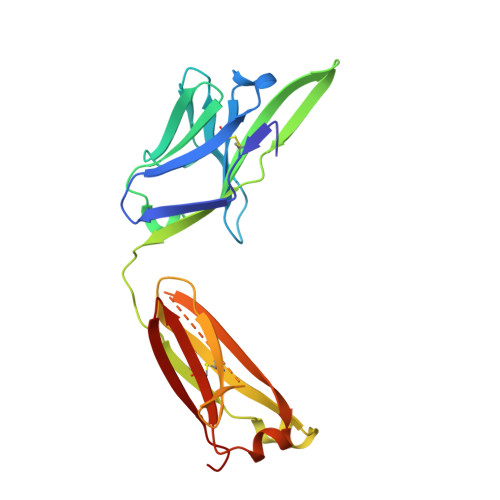
Disease-associated polyglutamine stretches in monomeric huntingtin adopt a compact structure.
Peters-Libeu, C., Miller, J., Rutenber, E., Newhouse, Y., Krishnan, P., Cheung, K., Hatters, D., Brooks, E., Widjaja, K., Tran, T., Mitra, S., Arrasate, M., Mosquera, L.A., Taylor, D., Weisgraber, K.H., Finkbeiner, S.(2012) J Mol Biology 421: 587-600
- PubMed: 22306738
- DOI: https://doi.org/10.1016/j.jmb.2012.01.034
- Primary Citation of Related Structures:
3S96, 4DCQ - PubMed Abstract:
Abnormal polyglutamine (polyQ) tracts are the only common feature in nine proteins that each cause a dominant neurodegenerative disorder. In Huntington's disease, tracts longer than 36 glutamines in the protein huntingtin (htt) cause degeneration. In situ, monoclonal antibody 3B5H10 binds to different htt fragments in neurons in proportion to their toxicity. Here, we determined the structure of 3B5H10 Fab to 1.9 Å resolution by X-ray crystallography. Modeling demonstrates that the paratope forms a groove suitable for binding two β-rich polyQ strands. Using small-angle X-ray scattering, we confirmed that the polyQ epitope recognized by 3B5H10 is a compact two-stranded hairpin within monomeric htt and is abundant in htt fragments unbound to antibody. Thus, disease-associated polyQ stretches preferentially adopt compact conformations. Since 3B5H10 binding predicts degeneration, this compact polyQ structure may be neurotoxic.
- Gladstone Institute of Neurological Disease, San Francisco, CA 94158, USA.
Organizational Affiliation: