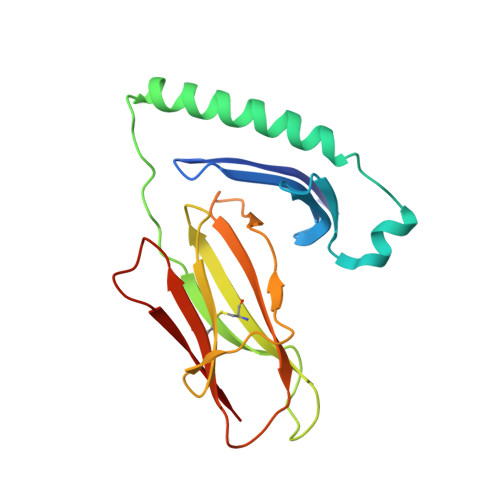
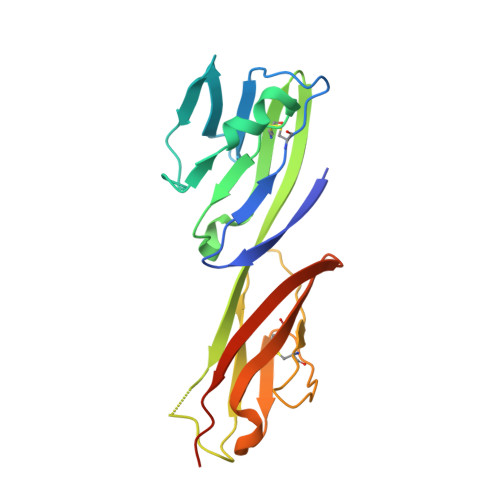Affinity maturation of human CD4 by yeast surface display and crystal structure of a CD4-HLA-DR1 complex.
Wang, X.X., Li, Y., Yin, Y., Mo, M., Wang, Q., Gao, W., Wang, L., Mariuzza, R.A.(2011) Proc Natl Acad Sci U S A 108: 15960-15965
- PubMed: 21900604
- DOI: https://doi.org/10.1073/pnas.1109438108
- Primary Citation of Related Structures:
3S4S, 3S5L - PubMed Abstract:
Helper T-cell activation generally requires the coreceptor CD4, which binds MHC class II molecules. A remarkable feature of the CD4-MHC class II interaction is its exceptionally low affinity, which ranges from K(D) = ∼200 μM to >2 mM. Investigating the biological role of the much lower affinity of this interaction than those of other cell-cell recognition molecules will require CD4 mutants with enhanced binding to MHC class II for testing in models of T-cell development. To this end, we used in vitro-directed evolution to increase the affinity of human CD4 for HLA-DR1. A mutant CD4 library was displayed on the surface of yeast and selected using HLA-DR1 tetramers or monomers, resulting in isolation of a CD4 clone containing 11 mutations. Reversion mutagenesis showed that most of the affinity increase derived from just two substitutions, Gln40Tyr and Thr45Trp. A CD4 variant bearing these mutations bound HLA-DR1 with K(D) = 8.8 μM, compared with >400 μM for wild-type CD4. To understand the basis for improved affinity, we determined the structure of this CD4 variant in complex with HLA-DR1 to 2.4 Å resolution. The structure provides an atomic-level description of the CD4-binding site on MHC class II and reveals how CD4 recognizes highly polymorphic HLA-DR, -DP, and -DQ molecules by targeting invariant residues in their α2 and β2 domains. In addition, the CD4 mutants reported here constitute unique tools for probing the influence of CD4 affinity on T-cell activation and development.
- W. M. Keck Laboratory for Structural Biology, University of Maryland Institute for Bioscience and Biotechnology Research, Rockville, MD 20850, USA.
Organizational Affiliation: