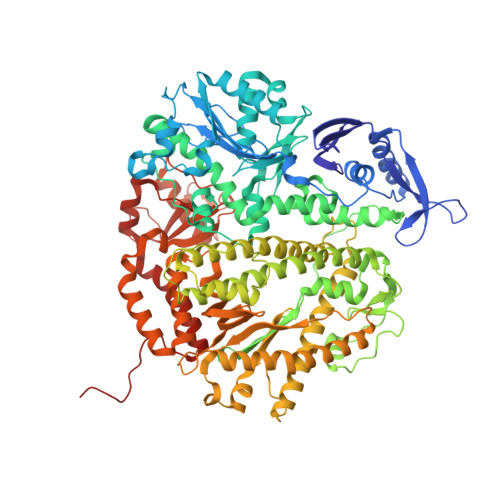
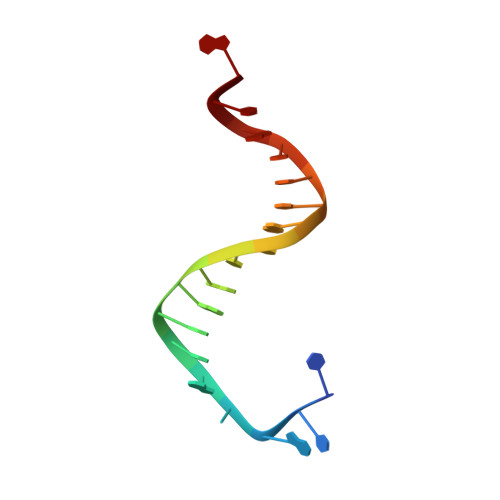
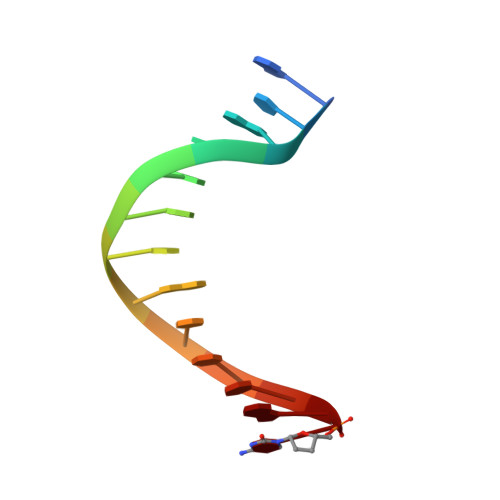
Hydrogen-Bonding Capability of a Templating Difluorotoluene Nucleotide Residue in an RB69 DNA Polymerase Ternary Complex.
Xia, S., Konigsberg, W.H., Wang, J.(2011) J Am Chem Soc 133: 10003-10005
- PubMed: 21667997
- DOI: https://doi.org/10.1021/ja2021735
- Primary Citation of Related Structures:
3QEP, 3RWU - PubMed Abstract:
Results obtained using 2,4-difluorotoluene nucleobase (dF) as a nonpolar thymine isostere by Kool and colleagues challenged the Watson-Crick dogma that hydrogen bonds between complementary bases are an absolute requirement for accurate DNA replication. Here, we report crystal structure of an RB69 DNA polymerase L561A/S565G/Y567A triple mutant ternary complex with a templating dF opposite dTTP at 1.8 Å-resolution. In this structure, direct hydrogen bonds were observed between: (i) dF and the incoming dTTP, (ii) dF and residue G568 of the polymerase, and (iii) dF and ordered water molecules surrounding the nascent base pair. Therefore, this structure provides evidence that a templating dF can form novel hydrogen bonds with the incoming dTTP and with the enzyme that differ from those formed with a templating dT.
- Department of Molecular Biophysics and Biochemistry, Yale University, New Haven, Connecticut 06520, USA.
Organizational Affiliation: