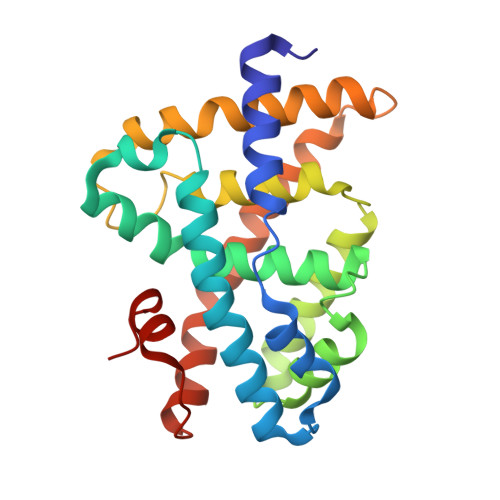
Conformationally constrained farnesoid X receptor (FXR) agonists: Alternative replacements of the stilbene.
Akwabi-Ameyaw, A., Caravella, J.A., Chen, L., Creech, K.L., Deaton, D.N., Madauss, K.P., Marr, H.B., Miller, A.B., Navas, F., Parks, D.J., Spearing, P.K., Todd, D., Williams, S.P., Wisely, G.B.(2011) Bioorg Med Chem Lett 21: 6154-6160
- PubMed: 21890356
- DOI: https://doi.org/10.1016/j.bmcl.2011.08.034
- Primary Citation of Related Structures:
3RUT, 3RUU, 3RVF - PubMed Abstract:
To further explore the optimum placement of the acid moiety in conformationally constrained analogs of GW 4064 1a, a series of stilbene replacements were prepared. The benzothiophene 1f and the indole 1g display the optimal orientation of the carboxylate for enhanced FXR agonist potency.
- Department of Medicinal Chemistry, GlaxoSmithKline, Research Triangle Park, NC 27709, USA.
Organizational Affiliation: