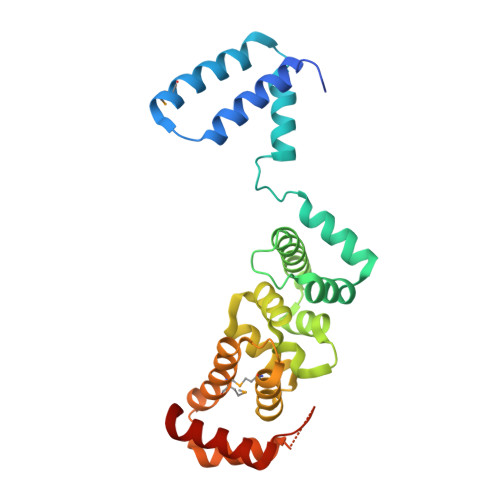
The Hyaloperonospora arabidopsidis ATR1 effector has distributed recognition surfaces and a structural subdomain conserved across oomycete species
Chou, S., Krasileva, K.V., Holton, J.M., Steinbrenner, A., Alber, T., Staskawicz, B.J.To be published.