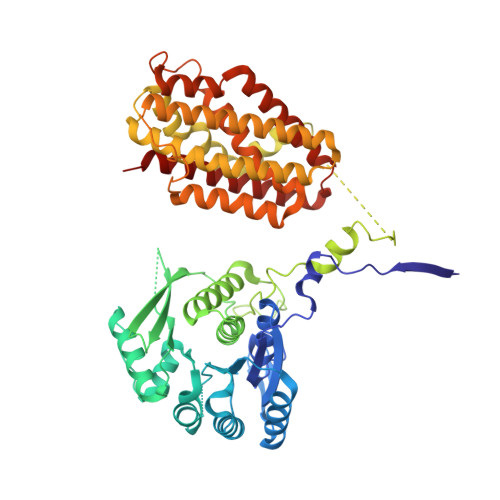
Structure of trifunctional THI20 from yeast.
French, J.B., Begley, T.P., Ealick, S.E.(2011) Acta Crystallogr D Biol Crystallogr 67: 784-791
- PubMed: 21904031
- DOI: https://doi.org/10.1107/S0907444911024814
- Primary Citation of Related Structures:
3RM5 - PubMed Abstract:
In a recently characterized thiamin-salvage pathway, thiamin-degradation products are hydrolyzed by thiaminase II, yielding 4-amino-5-hydroxymethyl-2-methylpyrimidine (HMP). This compound is an intermediate in thiamin biosynthesis that, once phosphorylated by an HMP kinase, can be used to synthesize thiamin monophosphate. Here, the crystal structure of Saccharomyces cerevisiae THI20, a trifunctional enzyme containing an N-terminal HMP kinase/HMP-P kinase (ThiD-like) domain and a C-terminal thiaminase II (TenA-like) domain, is presented. Comparison to structures of the monofunctional enzymes reveals that while the ThiD-like dimer observed in THI20 resembles other ThiD structures, the TenA-like domain, which is tetrameric in all previously reported structures, forms a dimer. Similarly, the active site of the ThiD-like domain of THI20 is highly similar to other known ThiD enzymes, while the TenA-like active site shows unique features compared with previously structurally characterized TenAs. In addition, a survey of known TenA structures revealed two structural classes, both of which have distinct conserved features. The TenA domain of THI20 possesses some features of both classes, consistent with its ability to hydrolyze both thiamin and the thiamin-degradation product 2-methyl-4-amino-5-aminomethylpyrimidine.
- Department of Chemistry and Chemical Biology, Cornell University, Ithaca, New York 14853-1301, USA.
Organizational Affiliation: