Mutational analysis of fungal family 11 xylanases on pH optimum determination
Fushinobu, S., Uno, T., Kitaoka, M., Hayashi, K., Matsuzawa, H., Wakagi, T.(2011) J Appl Glycosci (1999) 58: 107-114
Experimental Data Snapshot
Starting Model: experimental
View more details
wwPDB Validation 3D Report Full Report
(2011) J Appl Glycosci (1999) 58: 107-114
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Endo-1,4-beta-xylanase 3 | 185 | Aspergillus luchuensis | Mutation(s): 1 Gene Names: xynC EC: 3.2.1.8 | 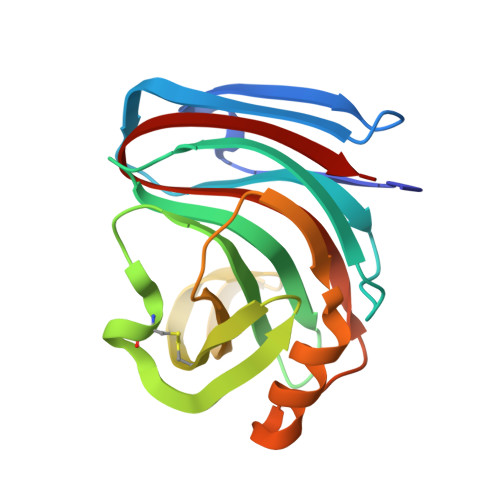 | |
UniProt | |||||
Find proteins for P33557 (Aspergillus kawachii (strain NBRC 4308)) Explore P33557 Go to UniProtKB: P33557 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P33557 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 62.159 | α = 90 |
| b = 62.159 | β = 90 |
| c = 113.536 | γ = 90 |
| Software Name | Purpose |
|---|---|
| DPS | data collection |
| REFMAC | refinement |
| MOSFLM | data reduction |
| SCALA | data scaling |