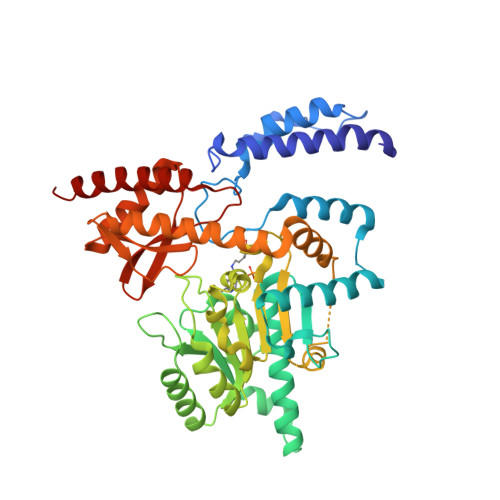
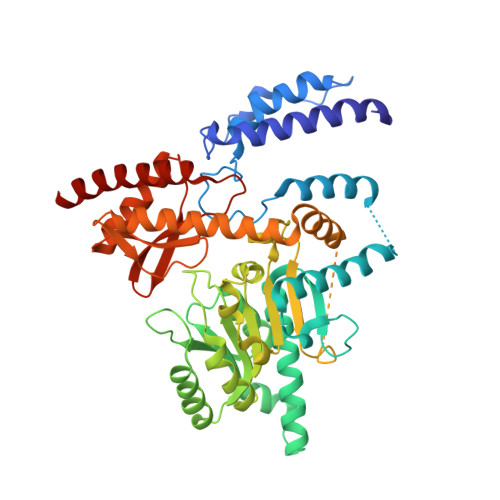
Open conformation of human DOPA decarboxylase reveals the mechanism of PLP addition to Group II decarboxylases.
Giardina, G., Montioli, R., Gianni, S., Cellini, B., Paiardini, A., Voltattorni, C.B., Cutruzzola, F.(2011) Proc Natl Acad Sci U S A 108: 20514-20519
- PubMed: 22143761
- DOI: https://doi.org/10.1073/pnas.1111456108
- Primary Citation of Related Structures:
3RBF, 3RBL, 3RCH - PubMed Abstract:
DOPA decarboxylase, the dimeric enzyme responsible for the synthesis of neurotransmitters dopamine and serotonin, is involved in severe neurological diseases such as Parkinson disease, schizophrenia, and depression. Binding of the pyridoxal-5'-phosphate (PLP) cofactor to the apoenzyme is thought to represent a central mechanism for the regulation of its activity. We solved the structure of the human apoenzyme and found it exists in an unexpected open conformation: compared to the pig kidney holoenzyme, the dimer subunits move 20 Å apart and the two active sites become solvent exposed. Moreover, by tuning the PLP concentration in the crystals, we obtained two more structures with different conformations of the active site. Analysis of three-dimensional data coupled to a kinetic study allows to identify the structural determinants of the open/close conformational change occurring upon PLP binding and thereby propose a model for the preferential degradation of the apoenzymes of Group II decarboxylases.
- Dipartimento di Scienze Biochimiche A. Rossi Fanelli, Sapienza Università di Roma, Piazzale Aldo Moro 5, 00185 Rome, Italy.
Organizational Affiliation: