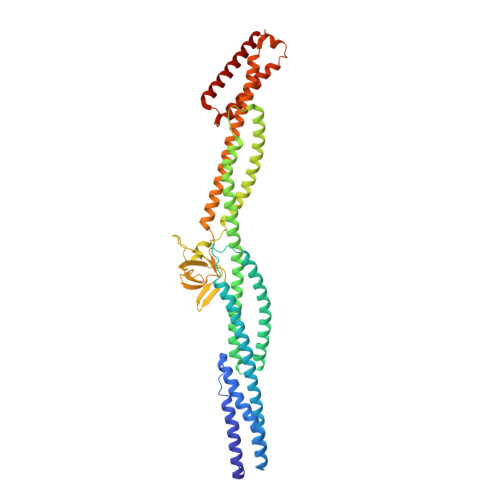
Crystal structure of a rigid four-spectrin-repeat fragment of the human desmoplakin plakin domain.
Choi, H.J., Weis, W.I.(2011) J Mol Biology 409: 800-812
- PubMed: 21536047
- DOI: https://doi.org/10.1016/j.jmb.2011.04.046
- Primary Citation of Related Structures:
3R6N - PubMed Abstract:
The plakin protein family serves to connect cell-cell and cell-matrix adhesion molecules to the intermediate filament cytoskeleton. Desmoplakin (DP) is an integral part of desmosomes, where it links desmosomal cadherins to the intermediate filaments. The 1056-amino-acid N-terminal region of DP contains a plakin domain common to members of the plakin family. Plakin domains contain multiple copies of spectrin repeats (SRs). We determined the crystal structure of a fragment of DP, residues 175-630, consisting of four SRs and an inserted SH3 domain. The four repeats form an elongated, rigid structure. The SH3 domain is present in a loop between two helices of an SR and interacts extensively with the preceding SR in a manner that appears to limit inter-repeat flexibility. The intimate intramolecular association of the SH3 domain with the preceding SR is also observed in plectin, another plakin protein, but not in α-spectrin, suggesting that the SH3 domain of plakins contributes to the stability and rigidity of this subfamily of SR-containing proteins.
- Department of Structural Biology, Stanford University School of Medicine, Stanford, CA 94305, USA.
Organizational Affiliation: