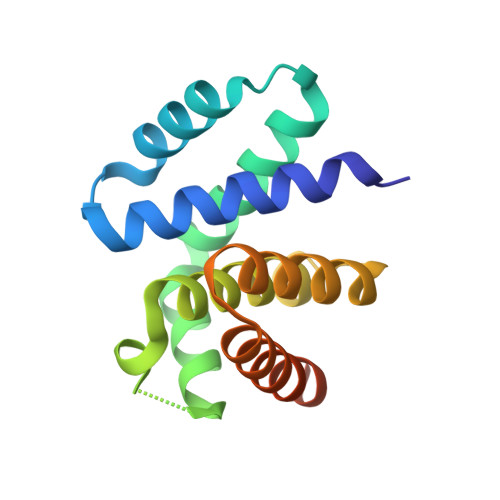
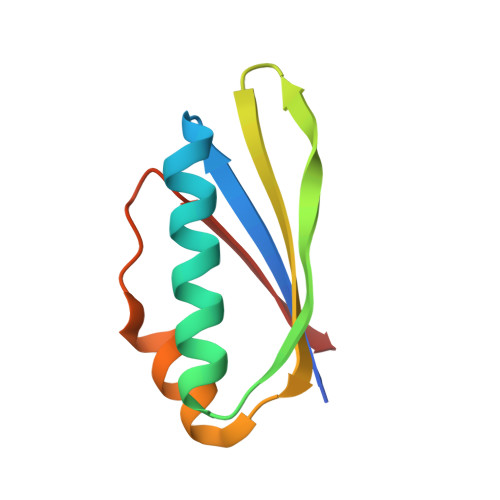
Structural basis for RNA recognition by NusB and NusE in the initiation of transcription antitermination.
Stagno, J.R., Altieri, A.S., Bubunenko, M., Tarasov, S.G., Li, J., Court, D.L., Byrd, R.A., Ji, X.(2011) Nucleic Acids Res 39: 7803-7815
- PubMed: 21652641
- DOI: https://doi.org/10.1093/nar/gkr418
- Primary Citation of Related Structures:
3R2C, 3R2D - PubMed Abstract:
Processive transcription antitermination requires the assembly of the complete antitermination complex, which is initiated by the formation of the ternary NusB-NusE-BoxA RNA complex. We have elucidated the crystal structure of this complex, demonstrating that the BoxA RNA is composed of 8 nt that are recognized by the NusB-NusE heterodimer. Functional biologic and biophysical data support the structural observations and establish the relative significance of key protein-protein and protein-RNA interactions. Further crystallographic investigation of a NusB-NusE-dsRNA complex reveals a heretofore unobserved dsRNA binding site contiguous with the BoxA binding site. We propose that the observed dsRNA represents BoxB RNA, as both single-stranded BoxA and double-stranded BoxB components are present in the classical lambda antitermination site. Combining these data with known interactions amongst antitermination factors suggests a specific model for the assembly of the complete antitermination complex.
- Macromolecular Crystallography Laboratory, National Cancer Institute, Frederick, MD 21702, USA.
Organizational Affiliation: