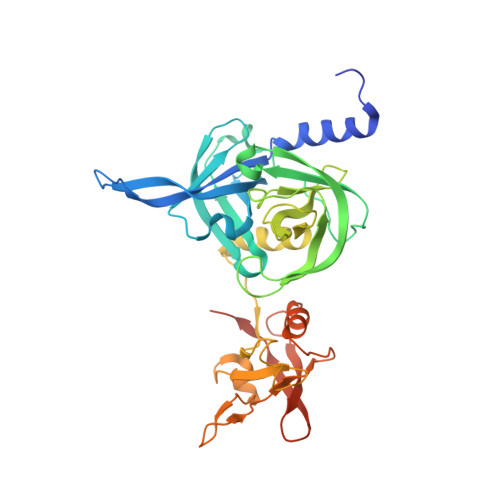
Structural adaptation of the plant protease Deg1 to repair photosystem II during light exposure.
Kley, J., Schmidt, B., Boyanov, B., Stolt-Bergner, P.C., Kirk, R., Ehrmann, M., Knopf, R.R., Naveh, L., Adam, Z., Clausen, T.(2011) Nat Struct Mol Biol 18: 728-731
- PubMed: 21532594
- DOI: https://doi.org/10.1038/nsmb.2055
- Primary Citation of Related Structures:
3QO6 - PubMed Abstract:
Deg1 is a chloroplastic protease involved in maintaining the photosynthetic machinery. Structural and biochemical analyses reveal that the inactive Deg1 monomer is transformed into the proteolytically active hexamer at acidic pH. The change in pH is sensed by His244, which upon protonation, repositions a specific helix to trigger oligomerization. This system ensures selective activation of Deg1 during daylight, when acidification of the thylakoid lumen occurs and photosynthetic proteins are damaged.
- Research Institute of Molecular Pathology (IMP), Vienna, Austria.
Organizational Affiliation: