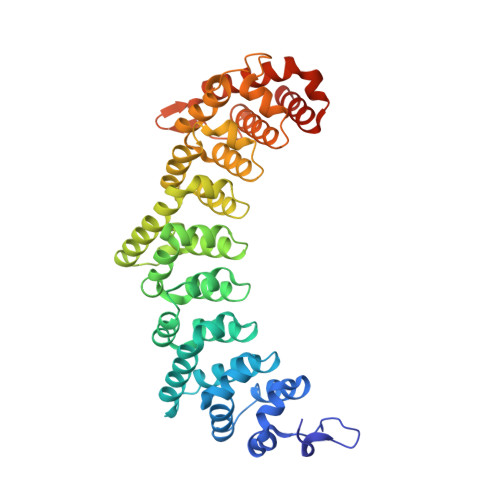
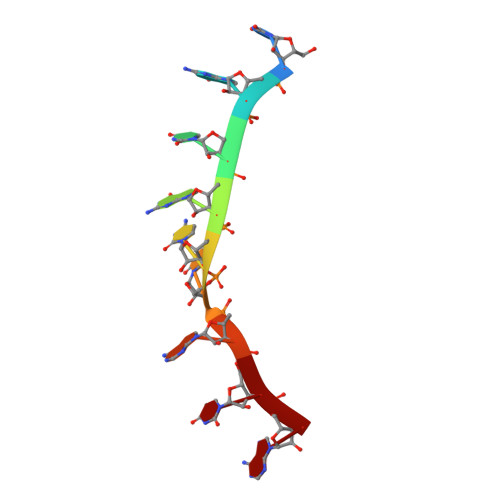
Stacking interactions in PUF-RNA complexes.
Koh, Y.Y., Wang, Y., Qiu, C., Opperman, L., Gross, L., Tanaka Hall, T.M., Wickens, M.(2011) RNA 17: 718-727
- PubMed: 21372189
- DOI: https://doi.org/10.1261/rna.2540311
- Primary Citation of Related Structures:
3QG9, 3QGB, 3QGC - PubMed Abstract:
Stacking interactions between amino acids and bases are common in RNA-protein interactions. Many proteins that regulate mRNAs interact with single-stranded RNA elements in the 3' UTR (3'-untranslated region) of their targets. PUF proteins are exemplary. Here we focus on complexes formed between a Caenorhabditis elegans PUF protein, FBF, and its cognate RNAs. Stacking interactions are particularly prominent and involve every RNA base in the recognition element. To assess the contribution of stacking interactions to formation of the RNA-protein complex, we combine in vivo selection experiments with site-directed mutagenesis, biochemistry, and structural analysis. Our results reveal that the identities of stacking amino acids in FBF affect both the affinity and specificity of the RNA-protein interaction. Substitutions in amino acid side chains can restrict or broaden RNA specificity. We conclude that the identities of stacking residues are important in achieving the natural specificities of PUF proteins. Similarly, in PUF proteins engineered to bind new RNA sequences, the identity of stacking residues may contribute to "target" versus "off-target" interactions, and thus be an important consideration in the design of proteins with new specificities.
- Department of Biochemistry, University of Wisconsin, Madison, Wisconsin 53706, USA.
Organizational Affiliation: