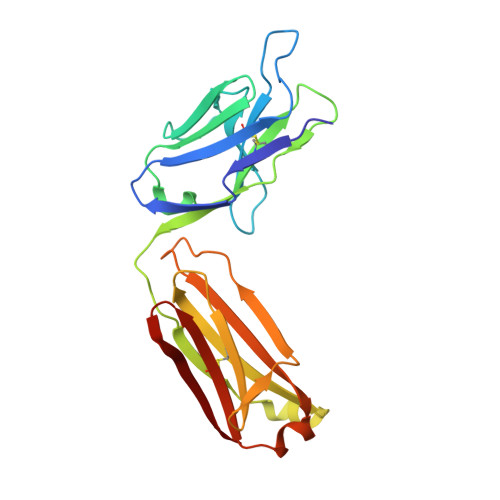
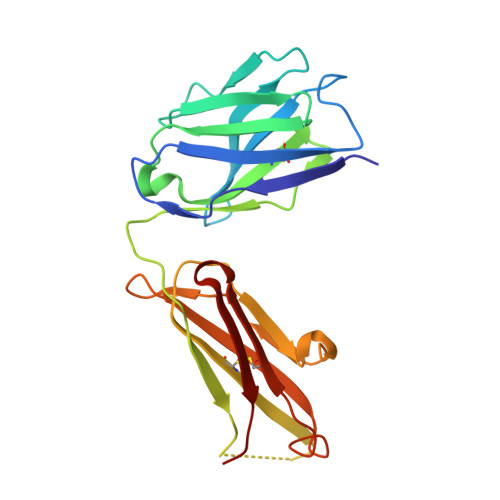Structural Basis for Ligand Recognition and Discrimination of a Quorum-quenching Antibody.
Kirchdoerfer, R.N., Garner, A.L., Flack, C.E., Mee, J.M., Horswill, A.R., Janda, K.D., Kaufmann, G.F., Wilson, I.A.(2011) J Biological Chem 286: 17351-17358
- PubMed: 21454495
- DOI: https://doi.org/10.1074/jbc.M111.231258
- Primary Citation of Related Structures:
3QG6, 3QG7 - PubMed Abstract:
In the postantibiotic era, available treatment options for severe bacterial infections caused by methicillin-resistant Staphylococcus aureus have become limited. Therefore, new and innovative approaches are needed to combat such life-threatening infections. Virulence factor expression in S. aureus is regulated in a cell density-dependent manner using "quorum sensing," which involves generation and secretion of autoinducing peptides (AIPs) into the surrounding environment to activate a bacterial sensor kinase at a particular threshold concentration. Mouse monoclonal antibody AP4-24H11 was shown previously to blunt quorum sensing-mediated changes in gene expression in vitro and protect mice from a lethal dose of S. aureus by sequestering the AIP signal. We have elucidated the crystal structure of the AP4-24H11 Fab in complex with AIP-4 at 2.5 Å resolution to determine its mechanism of ligand recognition. A key Glu(H95) provides much of the binding specificity through formation of hydrogen bonds with each of the four amide nitrogens in the AIP-4 macrocyclic ring. Importantly, these structural data give clues as to the interactions between the cognate staphylococcal AIP receptors AgrC and the AIPs, as AP4-24H11·AIP-4 binding recapitulates features that have been proposed for AgrC-AIP recognition. Additionally, these structural insights may enable the engineering of AIP cross-reactive antibodies or quorum quenching vaccines for use in active or passive immunotherapy for prevention or treatment of S. aureus infections.
- Department of Molecular Biology, The Scripps Research Institute, La Jolla, California 92037, USA.
Organizational Affiliation: