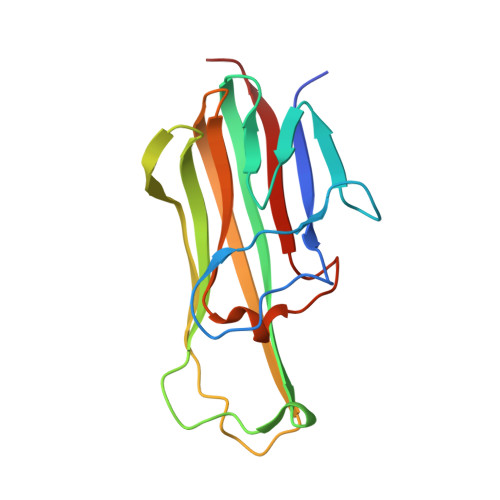
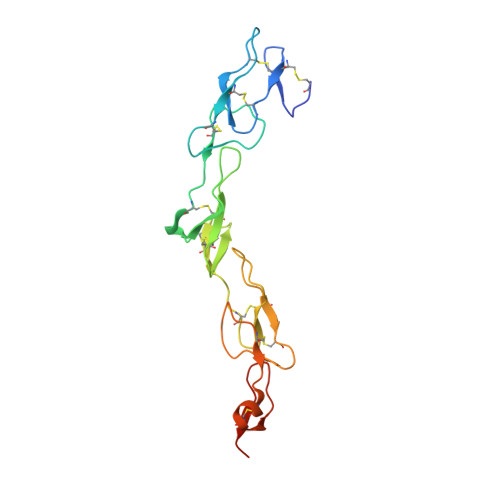
Structure-based development of a receptor activator of nuclear factor-kappaB ligand (RANKL) inhibitor peptide and molecular basis for osteopetrosis
Ta, H.M., Nguyen, G.T.T., Jin, H.M., Choi, J.K., Park, H., Kim, N.S., Hwang, H.Y., Kim, K.K.(2010) Proc Natl Acad Sci U S A 107: 20281-20286
- PubMed: 21059944
- DOI: https://doi.org/10.1073/pnas.1011686107
- Primary Citation of Related Structures:
3QBQ - PubMed Abstract:
The receptor activator of nuclear factor-κB (RANK) and its ligand RANKL, which belong to the tumor necrosis factor (TNF) receptor-ligand family, mediate osteoclastogenesis. The crystal structure of the RANKL ectodomain (eRANKL) in complex with the RANK ectodomain (eRANK) combined with biochemical assays of RANK mutants indicated that three RANK loops (Loop1, Loop2, and Loop3) bind to the interface of a trimeric eRANKL. Loop3 is particularly notable in that it is structurally distinctive from other TNF-family receptors and forms extensive contacts with RANKL. The disulfide bond (C125-C127) at the tip of Loop3 is important for determining the unique topology of Loop3, and docking E126 close to RANKL, which was supported by the inability of C127A or E126A mutants of RANK to bind to RANKL. Inhibitory activity of RANK mutants, which contain loops of osteoprotegerin (OPG), a soluble decoy receptor to RANKL, confirmed that OPG shares the similar binding mode with RANK and OPG. Loop3 plays a key role in RANKL binding. Peptide inhibitors designed to mimic Loop3 blocked the RANKL-induced differentiation of osteoclast precursors, suggesting that they could be developed as therapeutic agents for the treatment of osteoporosis and bone-related diseases. Furthermore, some of the RANK mutations associated with autosomal recessive osteopetrosis (ARO) resulted in reduced RANKL-binding activity and failure to induce osteoclastogenesis. These results, together with structural interpretation of eRANK-eRANKL interaction, provided molecular understanding for pathogenesis of ARO.
- Department of Molecular Cell Biology, Samsung Biomedical Research Institute, Sungkyunkwan University School of Medicine, Suwon 440-746, Korea.
Organizational Affiliation: