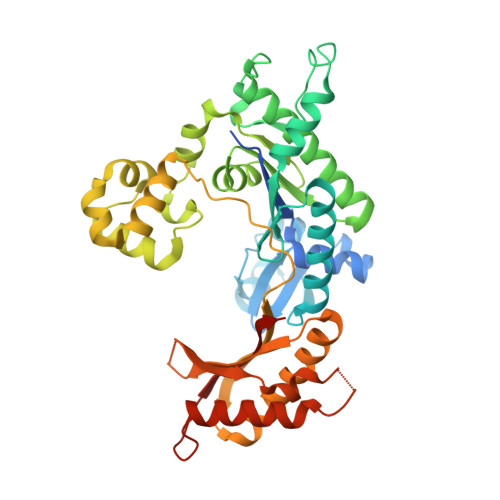
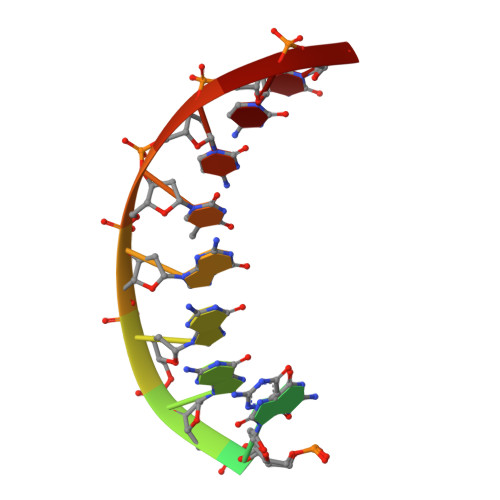
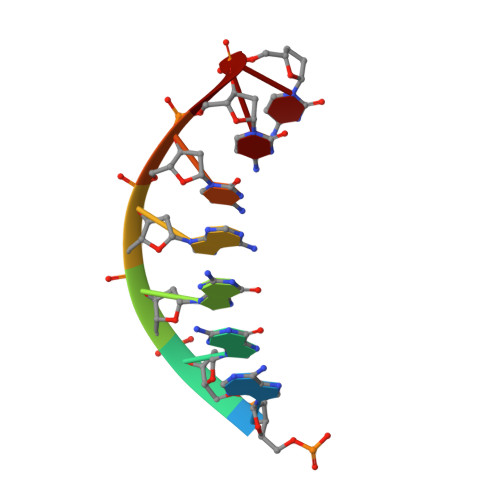
Unique active site promotes error-free replication opposite an 8-oxo-guanine lesion by human DNA polymerase iota.
Kirouac, K.N., Ling, H.(2011) Proc Natl Acad Sci U S A 108: 3210-3215
- PubMed: 21300901
- DOI: https://doi.org/10.1073/pnas.1013909108
- Primary Citation of Related Structures:
3Q8P, 3Q8Q, 3Q8R - PubMed Abstract:
The 8-oxo-guanine (8-oxo-G) lesion is the most abundant and mutagenic oxidative DNA damage existing in the genome. Due to its dual coding nature, 8-oxo-G causes most DNA polymerases to misincorporate adenine. Human Y-family DNA polymerase iota (polι) preferentially incorporates the correct cytosine nucleotide opposite 8-oxo-G. This unique specificity may contribute to polι's biological role in cellular protection against oxidative stress. However, the structural basis of this preferential cytosine incorporation is currently unknown. Here we present four crystal structures of polι in complex with DNA containing an 8-oxo-G lesion, paired with correct dCTP or incorrect dATP, dGTP, and dTTP nucleotides. An exceptionally narrow polι active site restricts the purine bases in a syn conformation, which prevents the dual coding properties of 8-oxo-G by inhibiting syn/anti conformational equilibrium. More importantly, the 8-oxo-G base in a syn conformation is not mutagenic in polι because its Hoogsteen edge does not form a stable base pair with dATP in the narrow active site. Instead, the syn 8-oxo-G template base forms the most stable replicating base pair with correct dCTP due to its small pyrimidine base size and enhanced hydrogen bonding with the Hoogsteen edge of 8-oxo-G. In combination with site directed mutagenesis, we show that Gln59 in the finger domain specifically interacts with the additional O(8) atom of the lesion base, which influences nucleotide selection, enzymatic efficiency, and replication stalling at the lesion site. Our work provides the structural mechanism of high-fidelity 8-oxo-G replication by a human DNA polymerase.
- Department of Biochemistry, University of Western Ontario, London, ON, Canada N6A 5C1.
Organizational Affiliation: