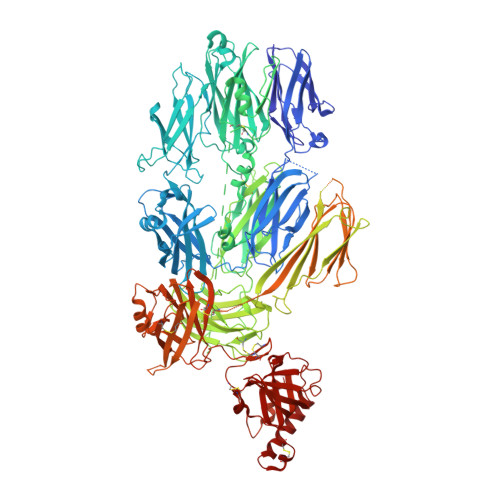
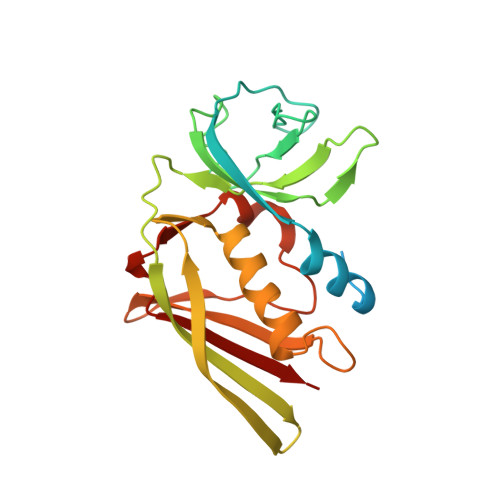
Substrate recognition by complement convertases revealed in the C5-cobra venom factor complex.
Laursen, N.S., Andersen, K.R., Braren, I., Spillner, E., Sottrup-Jensen, L., Andersen, G.R.(2011) EMBO J 30: 606-616
- PubMed: 21217642
- DOI: https://doi.org/10.1038/emboj.2010.341
- Primary Citation of Related Structures:
3PRX, 3PVM - PubMed Abstract:
Complement acts as a danger-sensing system in the innate immune system, and its activation initiates a strong inflammatory response and cleavage of the proteins C3 and C5 by proteolytic enzymes, the convertases. These contain a non-catalytic substrate contacting subunit (C3b or C4b) in complex with a protease subunit (Bb or C2a). We determined the crystal structures of the C3b homologue cobra venom factor (CVF) in complex with C5, and in complex with C5 and the inhibitor SSL7 at 4.3 Å resolution. The structures reveal a parallel two-point attachment between C5 and CVF, where the presence of SSL7 only slightly affects the C5-CVF interface, explaining the IgA dependence for SSL7-mediated inhibition of C5 cleavage. CVF functions as a relatively rigid binding scaffold inducing a conformational change in C5, which positions its cleavage site in proximity to the serine protease Bb. A general model for substrate recognition by the convertases is presented based on the C5-CVF and C3b-Bb-SCIN structures. Prior knowledge concerning interactions between the endogenous convertases and their substrates is rationalized by this model.
- Department of Molecular Biology, Aarhus University, Aarhus, Denmark.
Organizational Affiliation: