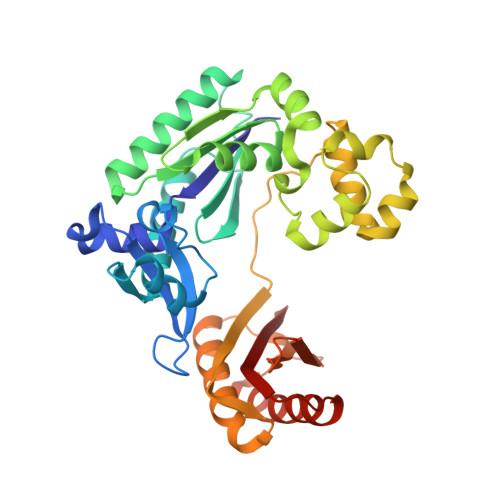
Structural mechanism of ribonucleotide discrimination by a Y-family DNA polymerase.
Kirouac, K.N., Suo, Z., Ling, H.(2011) J Mol Biology 407: 382-390
- PubMed: 21295588
- DOI: https://doi.org/10.1016/j.jmb.2011.01.037
- Primary Citation of Related Structures:
3PR4, 3PR5 - PubMed Abstract:
The ability of DNA polymerases to differentiate between ribonucleotides and deoxribonucleotides is fundamental to the accurate replication and maintenance of an organism's genome. The active sites of Y-family DNA polymerases are highly solvent accessible, yet these enzymes still maintain a high selectivity towards deoxyribonucleotides. Here, we biochemically demonstrate that a single active-site mutation (Y12A) in Dpo4, a model Y-family DNA polymerase, causes both a dramatic loss of ribonucleotide discrimination and a decrease in nucleotide incorporation efficiency. We also determined two ternary crystal structures of the Dpo4 Y12A mutant incorporating either dATP or ATP nucleotides opposite a template dT base. Interestingly, both dATP and ATP were hydrolyzed to dADP and ADP, respectively. In addition, the dADP and ADP molecules adopt a similar conformation and position at the polymerase active site to a ddADP molecule in the ternary crystal structure of wild-type Dpo4. The Y12A mutant loses stacking interactions with the deoxyribose of dNTP, which destabilizes the binding of incoming nucleotides. The mutation also opens a space to accommodate the 2'-OH group of the ribose of NTP in the polymerase active site. The structural change leads to the reduction in deoxynucleotide incorporation efficiency and allows ribonucleotide incorporation.
- Department of Biochemistry, University of Western Ontario, London, Ontario, Canada N6A 5C1.
Organizational Affiliation: