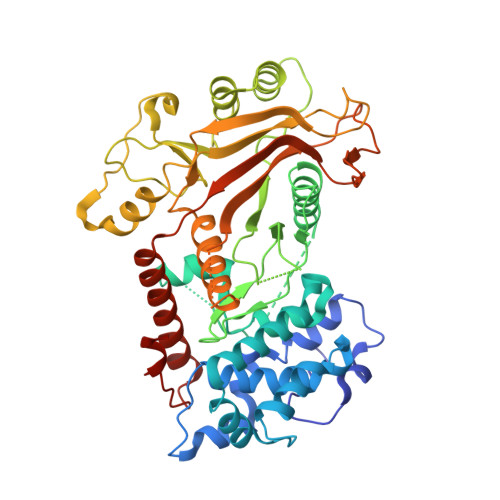
Crystal structure of a KSHV-SOX-DNA complex: insights into the molecular mechanisms underlying DNase activity and host shutoff
Bagneris, C., Briggs, L.C., Savva, R., Ebrahimi, B., Barrett, T.E.(2011) Nucleic Acids Res 39: 5744-5756
- PubMed: 21421561
- DOI: https://doi.org/10.1093/nar/gkr111
- Primary Citation of Related Structures:
3POV - PubMed Abstract:
The early lytic phase of Kaposi's sarcoma herpesvirus infection is characterized by viral replication and the global degradation (shutoff) of host mRNA. Key to both activities is the virally encoded alkaline exonuclease KSHV SOX. While the DNase activity of KSHV SOX is required for the resolution of viral genomic DNA as a precursor to encapsidation, its exact involvement in host shutoff remains to be determined. We present the first crystal structure of a KSHV SOX-DNA complex that has illuminated the catalytic mechanism underpinning both its endo and exonuclease activities. We further illustrate that KSHV SOX, similar to its Epstein-Barr virus homologue, has an intrinsic RNase activity in vitro that although an element of host shutoff, cannot solely account for the phenomenon.
- Institute of Structural and Molecular Biology, Crystallography, Department of Biological Sciences, Birkbeck College, Malet Street, London WC1E 7HX , UK.
Organizational Affiliation: