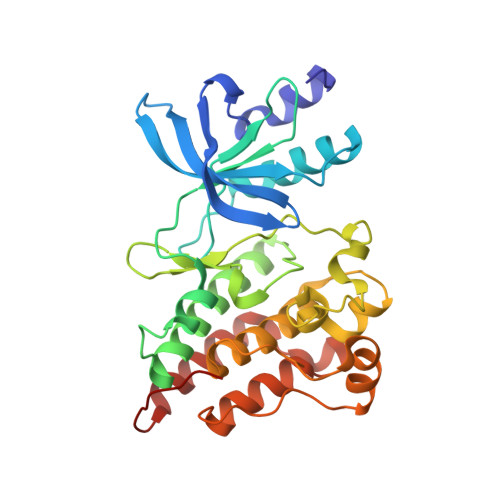
The Crystal Structure of a Constitutively Active Mutant RON Kinase Suggests an Intramolecular Autophosphorylation Hypothesis
Wang, J., Steinbacher, S., Augustin, M., Schreiner, P., Epstein, D., Mulvihill, M.J., Crew, A.P.(2010) Biochemistry 49: 7972-7974
- PubMed: 20726546
- DOI: https://doi.org/10.1021/bi100409w
- Primary Citation of Related Structures:
3PLS - PubMed Abstract:
A complex of RON(M1254T) with AMP-PNP and Mg(2+) reveals a substratelike positioning of Tyr1238 as well as likely catalysis-competent placement of the AMP-PNP and Mg(2+) components and indicates a tendency for cis phosphorylation. The structure shows how the oncogenic mutation may cause the constitutive activation and suggests a mechanistic hypothesis for the autophosphorylation of receptor tyrosine kinases.
- OSI Pharmaceuticals, Inc., 1 Bioscience Park Drive, Farmingdale, New York 11735, USA. jwang@osip.com
Organizational Affiliation: