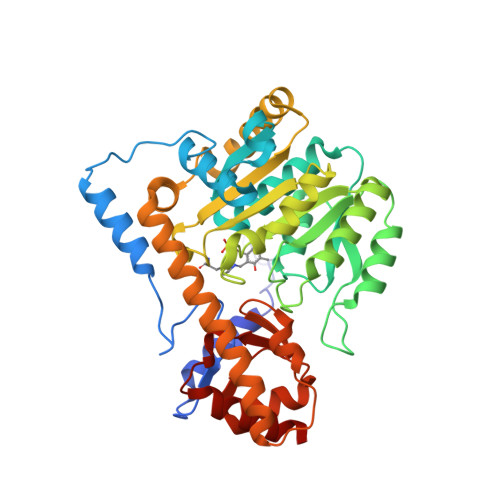
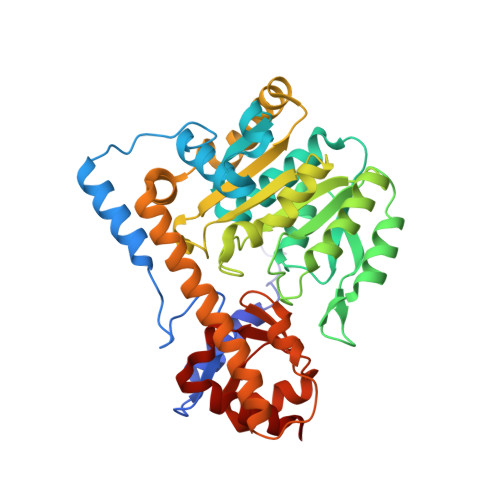
Biochemical and structural characterization of mouse mitochondrial aspartate aminotransferase, a newly identified kynurenine aminotransferase-IV.
Han, Q., Robinson, H., Cai, T., Tagle, D.A., Li, J.(2011) Biosci Rep 31: 323-332
- PubMed: 20977429
- DOI: https://doi.org/10.1042/BSR20100117
- Primary Citation of Related Structures:
3PD6, 3PDB - PubMed Abstract:
Mammalian mAspAT (mitochondrial aspartate aminotransferase) is recently reported to have KAT (kynurenine aminotransferase) activity and plays a role in the biosynthesis of KYNA (kynurenic acid) in rat, mouse and human brains. This study concerns the biochemical and structural characterization of mouse mAspAT. In this study, mouse mAspAT cDNA was amplified from mouse brain first stand cDNA and its recombinant protein was expressed in an Escherichia coli expression system. Sixteen oxo acids were tested for the co-substrate specificity of mouse mAspAT and 14 of them were shown to be capable of serving as co-substrates for the enzyme. Structural analysis of mAspAT by macromolecular crystallography revealed that the cofactor-binding residues of mAspAT are similar to those of other KATs. The substrate-binding residues of mAspAT are slightly different from those of other KATs. Our results provide a biochemical and structural basis towards understanding the overall physiological role of mAspAT in vivo and insight into controlling the levels of endogenous KYNA through modulation of the enzyme in the mouse brain.
- Department of Biochemistry, Virginia Tech, Blacksburg, VA 24061, USA.
Organizational Affiliation: