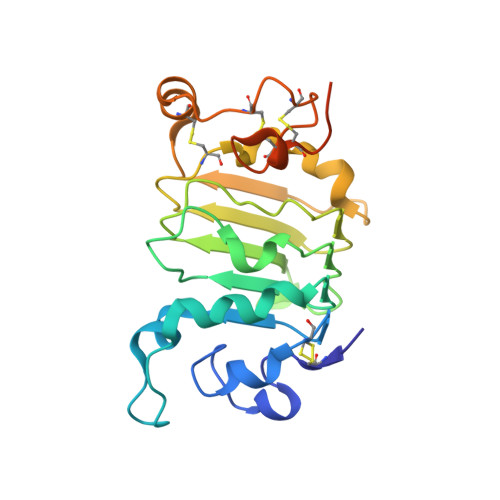
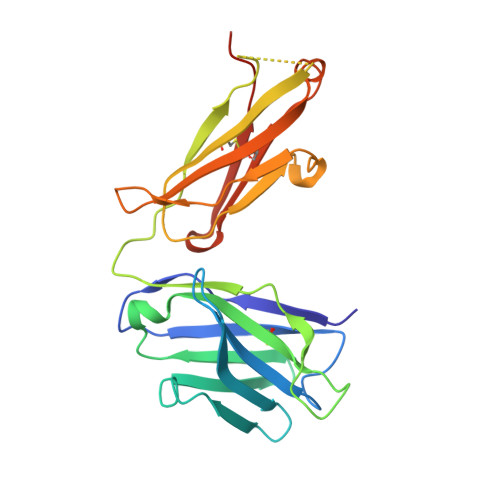
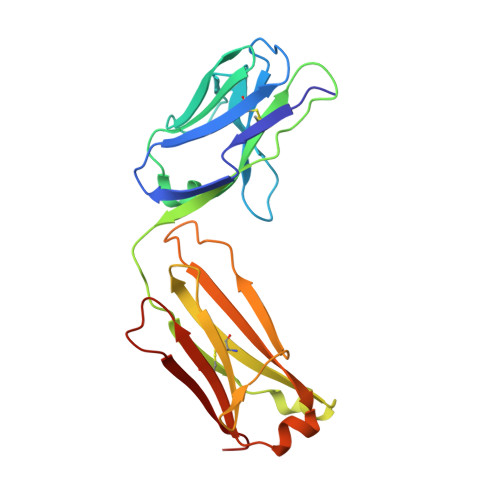
A two-in-one antibody against HER3 and EGFR has superior inhibitory activity compared with monospecific antibodies.
Schaefer, G., Haber, L., Crocker, L.M., Shia, S., Shao, L., Dowbenko, D., Totpal, K., Wong, A., Lee, C.V., Stawicki, S., Clark, R., Fields, C., Lewis Phillips, G.D., Prell, R.A., Danilenko, D.M., Franke, Y., Stephan, J.P., Hwang, J., Wu, Y., Bostrom, J., Sliwkowski, M.X., Fuh, G., Eigenbrot, C.(2011) Cancer Cell 20: 472-486
- PubMed: 22014573
- DOI: https://doi.org/10.1016/j.ccr.2011.09.003
- Primary Citation of Related Structures:
3P0V, 3P0Y, 3P11 - PubMed Abstract:
Extensive crosstalk among ErbB/HER receptors suggests that blocking signaling from more than one family member may be essential to effectively treat cancer and limit drug resistance. We generated a conventional IgG molecule MEHD7945A with dual HER3/EGFR specificity by phage display engineering and used structural and mutational studies to understand how a single antigen recognition surface binds two epitopes with high affinity. As a human IgG1, MEHD7945A exhibited dual action by inhibiting EGFR- and HER3-mediated signaling in vitro and in vivo and the ability to engage immune effector functions. Compared with monospecific anti-HER antibodies, MEHD7945A was more broadly efficacious in multiple tumor models, showing that combined inhibition of EGFR and HER3 with a single antibody is beneficial.
- Department of Research Oncology, Genentech, Inc, South San Francisco, CA 94080, USA.
Organizational Affiliation: