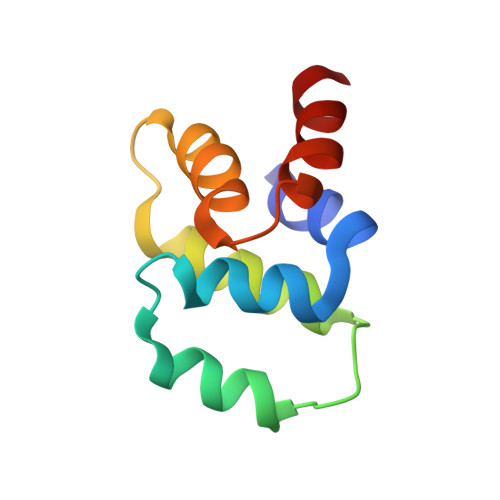
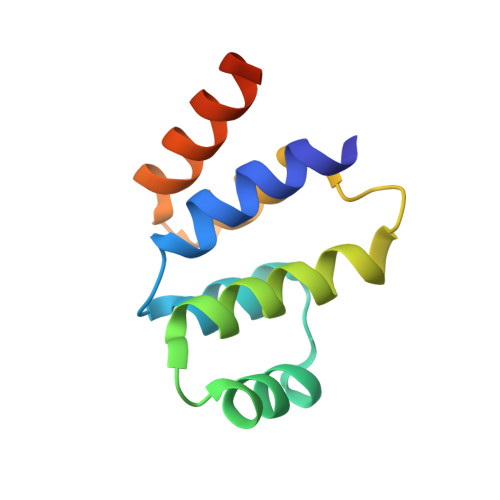
The Fas-FADD death domain complex structure reveals the basis of DISC assembly and disease mutations.
Wang, L., Yang, J.K., Kabaleeswaran, V., Rice, A.J., Cruz, A.C., Park, A.Y., Yin, Q., Damko, E., Jang, S.B., Raunser, S., Robinson, C.V., Siegel, R.M., Walz, T., Wu, H.(2010) Nat Struct Mol Biol 17: 1324-1329
- PubMed: 20935634
- DOI: https://doi.org/10.1038/nsmb.1920
- Primary Citation of Related Structures:
3OQ9 - PubMed Abstract:
The death-inducing signaling complex (DISC) formed by the death receptor Fas, the adaptor protein FADD and caspase-8 mediates the extrinsic apoptotic program. Mutations in Fas that disrupt the DISC cause autoimmune lymphoproliferative syndrome (ALPS). Here we show that the Fas-FADD death domain (DD) complex forms an asymmetric oligomeric structure composed of 5-7 Fas DD and 5 FADD DD, whose interfaces harbor ALPS-associated mutations. Structure-based mutations disrupt the Fas-FADD interaction in vitro and in living cells; the severity of a mutation correlates with the number of occurrences of a particular interaction in the structure. The highly oligomeric structure explains the requirement for hexameric or membrane-bound FasL in Fas signaling. It also predicts strong dominant negative effects from Fas mutations, which are confirmed by signaling assays. The structure optimally positions the FADD death effector domain (DED) to interact with the caspase-8 DED for caspase recruitment and higher-order aggregation.
- Department of Biochemistry, Weill Cornell Medical College, New York, New York, USA.
Organizational Affiliation: