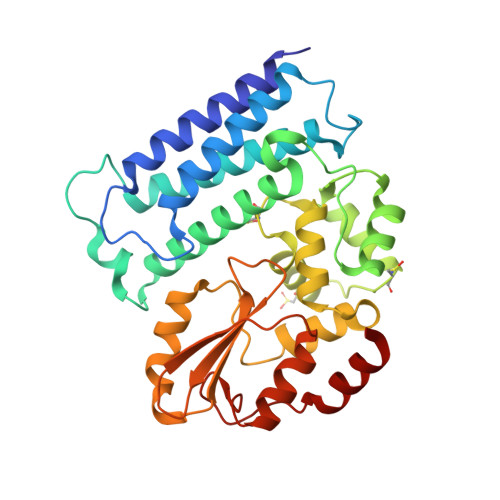
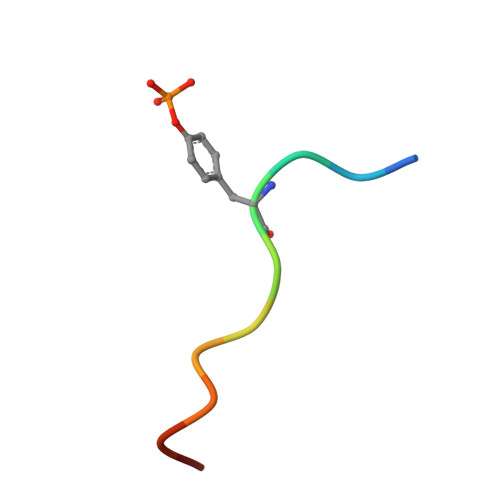
Crystal structure of Cbl-c (Cbl-3) TKB domain in complex with EGFR pY1069 peptide
Chaikuad, A., Guo, K., Cooper, C.D.O., Ayinampudi, V., Krojer, T., Ugochukwu, E., Muniz, J.R.C., Vollmar, M., Canning, P., von Delft, F., Arrowsmith, C.H., Weigelt, J., Edwards, A.M., Bountra, C., Bullock, A.To be published.