Structural basis of chemokine sequestration by CrmD, a poxvirus-encoded tumor necrosis factor receptor
Xue, X.G., Lu, Q.Y., Wei, H., Wang, D.L., Chen, D.W., He, G.J., Huang, L., Wang, H.Z., Wang, X.Q.(2011) PLoS Pathog 7: e1002162-e1002162
- PubMed: 21829356
- DOI: https://doi.org/10.1371/journal.ppat.1002162
- Primary Citation of Related Structures:
3ON9, 3ONA - PubMed Abstract:
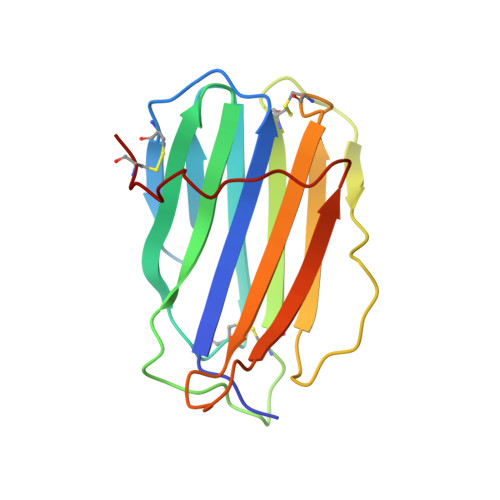
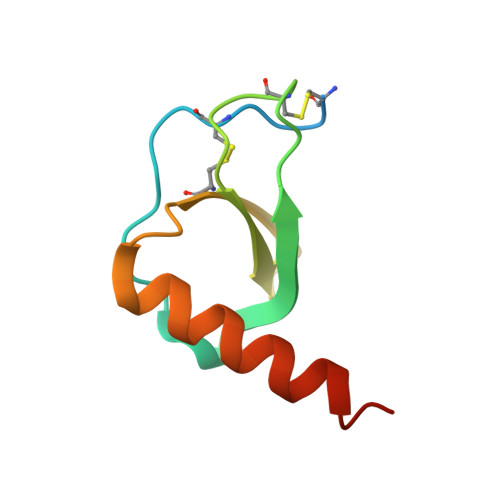
Pathogens have evolved sophisticated mechanisms to evade detection and destruction by the host immune system. Large DNA viruses encode homologues of chemokines and their receptors, as well as chemokine-binding proteins (CKBPs) to modulate the chemokine network in host response. The SECRET domain (smallpox virus-encoded chemokine receptor) represents a new family of viral CKBPs that binds a subset of chemokines from different classes to inhibit their activities, either independently or fused with viral tumor necrosis factor receptors (vTNFRs). Here we present the crystal structures of the SECRET domain of vTNFR CrmD encoded by ectromelia virus and its complex with chemokine CX3CL1. The SECRET domain adopts a β-sandwich fold and utilizes its β-sheet I surface to interact with CX3CL1, representing a new chemokine-binding manner of viral CKBPs. Structure-based mutagenesis and biochemical analysis identified important basic residues in the 40s loop of CX3CL1 for the interaction. Mutation of corresponding acidic residues in the SECRET domain also affected the binding for other chemokines, indicating that the SECRET domain binds different chemokines in a similar manner. We further showed that heparin inhibited the binding of CX3CL1 by the SECRET domain and the SECRET domain inhibited RAW264.7 cell migration induced by CX3CL1. These results together shed light on the structural basis for the SECRET domain to inhibit chemokine activities by interfering with both chemokine-GAG and chemokine-receptor interactions.
- Center for Structural Biology, School of Life Sciences, Ministry of Education Key Laboratory of Protein Sciences, Tsinghua University, Beijing, People's Republic of China.
Organizational Affiliation: