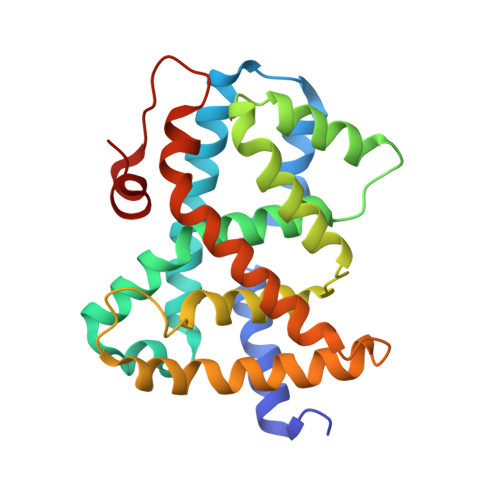
Optimization of a novel class of benzimidazole-based farnesoid X receptor (FXR) agonists to improve physicochemical and ADME properties
Richter, H.G.F., Benson, G.M., Bleicher, K.H., Blum, D., Chaput, E., Clemann, N., Feng, S., Gardes, C., Grether, U., Hartman, P., Kuhn, B., Martin, R.E., Plancher, J.M., Rudolph, M.G., Schuler, F., Taylor, S.(2011) Bioorg Med Chem Lett 21: 1134-1140
- PubMed: 21269824
- DOI: https://doi.org/10.1016/j.bmcl.2010.12.123
- Primary Citation of Related Structures:
3OLF, 3OMK, 3OMM, 3OOF, 3OOK - PubMed Abstract:
Structure-guided lead optimization of recently described benzimidazolyl acetamides addressed the key liabilities of the previous lead compound 1. These efforts culminated in the discovery of 4-{(S)-2-[2-(4-chloro-phenyl)-5,6-difluoro-benzoimidazol-1-yl]-2-cyclohexyl-acetylamino}-3-fluoro-benzoic acid 7g, a highly potent and selective FXR agonist with excellent physicochemical and ADME properties and potent lipid lowering activity after oral administration to LDL receptor deficient mice.
- F. Hoffmann-La Roche Ltd, Pharmaceutical Research, Grenzacherstrasse, CH-4070 Basel, Switzerland. hans.richter@roche.com
Organizational Affiliation: