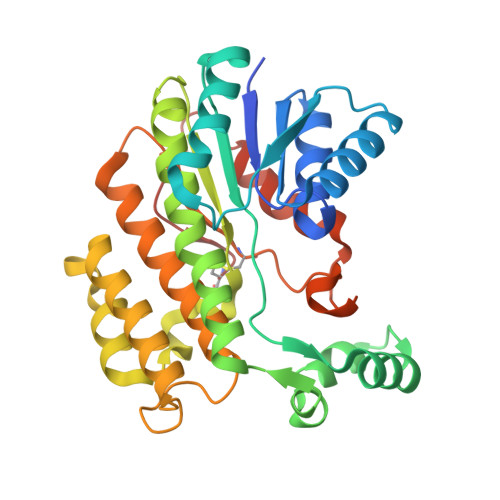
The atomic structure of salutaridine reductase from the opium poppy Papaver somniferum.
Higashi, Y., Kutchan, T.M., Smith, T.J.(2010) J Biological Chem 66: 163-166
- PubMed: 21169353
- DOI: https://doi.org/10.1074/jbc.M110.168633
- Primary Citation of Related Structures:
3O26 - PubMed Abstract:
The opium poppy (Papaver somniferum L.) is one of the oldest known medicinal plants. In the biosynthetic pathway for morphine and codeine, salutaridine is reduced to salutaridinol by salutaridine reductase (SalR; EC 1.1.1.248) using NADPH as coenzyme. Here, we report the atomic structure of SalR to a resolution of ∼1.9 Å in the presence of NADPH. The core structure is highly homologous to other members of the short chain dehydrogenase/reductase family. The major difference is that the nicotinamide moiety and the substrate-binding pocket are covered by a loop (residues 265-279), on top of which lies a large "flap"-like domain (residues 105-140). This configuration appears to be a combination of the two common structural themes found in other members of the short chain dehydrogenase/reductase family. Previous modeling studies suggested that substrate inhibition is due to mutually exclusive productive and nonproductive modes of substrate binding in the active site. This model was tested via site-directed mutagenesis, and a number of these mutations abrogated substrate inhibition. However, the atomic structure of SalR shows that these mutated residues are instead distributed over a wide area of the enzyme, and many are not in the active site. To explain how residues distal to the active site might affect catalysis, a model is presented whereby SalR may undergo significant conformational changes during catalytic turnover.
- Donald Danforth Plant Science Center, St. Louis, Missouri 63132, USA.
Organizational Affiliation: