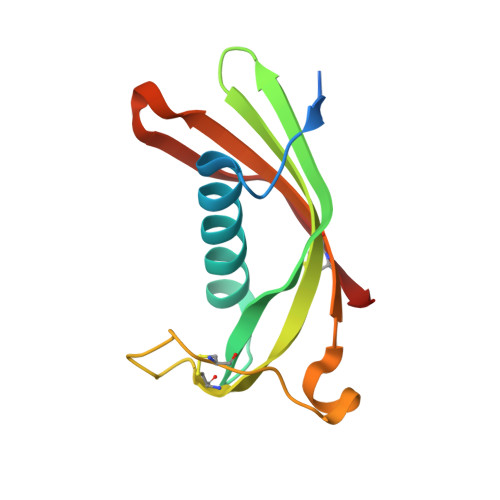
Hinge-loop mutation can be used to control 3D domain swapping and amyloidogenesis of human cystatin C.
Orlikowska, M., Jankowska, E., Kolodziejczyk, R., Jaskolski, M., Szymanska, A.(2011) J Struct Biol 173: 406-413
- PubMed: 21074623
- DOI: https://doi.org/10.1016/j.jsb.2010.11.009
- Primary Citation of Related Structures:
3NX0 - PubMed Abstract:
Cystatins are natural inhibitors of cysteine proteases, enzymes that are widely distributed in animals, plants, and microorganisms. Human cystatin C (hCC) has been also recognized as an aggregating protein directly involved in the formation of pathological amyloid fibrils, and these amyloidogenic properties greatly increase in a naturally occurring L68Q hCC variant. For a long time only dimeric structure of wild-type hCC has been known. The dimer is created through 3D domain swapping process, in which two parts of the cystatin structure become separated from each other and next exchanged between two molecules. Important role in the domain swapping plays the L1 loop, which connects the exchanging segments and, upon dimerization, transforms from a β-turn into a part of a long β-strand. In the very recently published first monomeric structure of human cystatin C (hCC-stab1), dimerization was abrogated due to clasping of the β-strands from the swapping domains by an engineered disulfide bridge. We have designed and constructed another mutated cystatin C with the smallest possible structural intervention, that is a single-point mutation replacing hydrophobic V57 from the L1 loop by polar asparagine, known as a stabilizer of a β-turn motif. V57N hCC mutant occurred to be stable in its monomeric form and crystallized as a monomer, revealing typical cystatin fold with a five-stranded antiparallel β-sheet wrapped around an α-helix. Here we report a 2.04 Å resolution crystal structure of V57N hCC and discuss the architecture of the protein in comparison to chicken cystatin, hCC-stab1 and dimeric hCC.
- Department of Medicinal Chemistry, Faculty of Chemistry, University of Gdańsk, Gdańsk, Poland.
Organizational Affiliation: