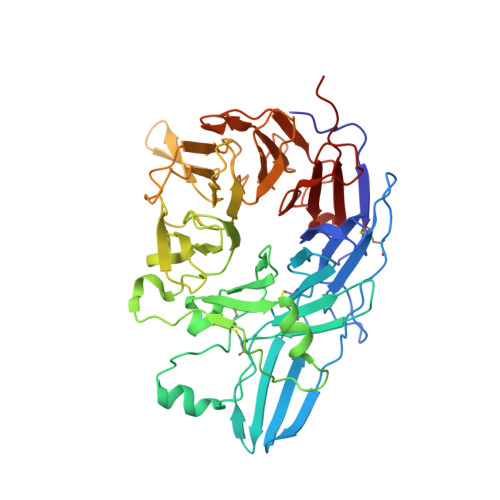
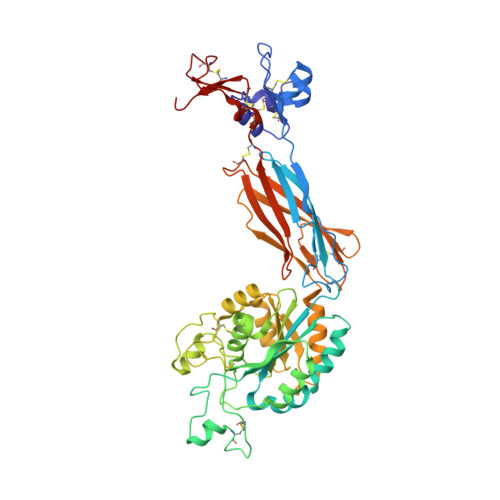
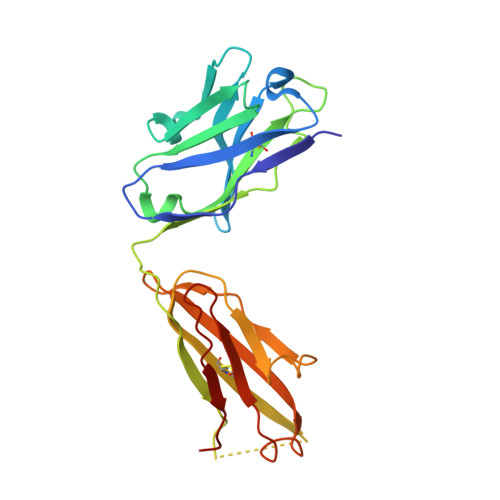
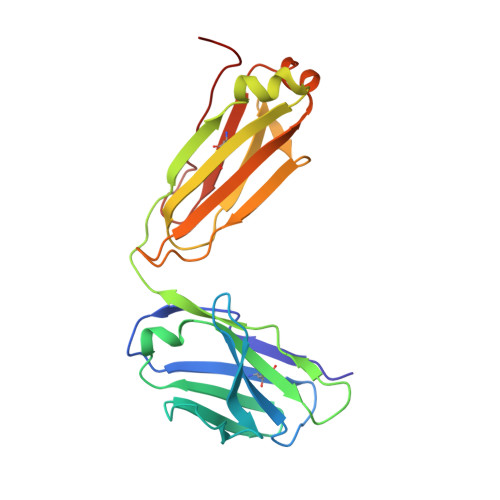
Closed headpiece of integrin {alpha}IIb{beta}3 and its complex with an {alpha}IIb{beta}3-specific antagonist that does not induce opening.
Zhu, J.H., Zhu, J.Q., Negri, A., Provasi, D., Filizola, M., Coller, B.S., Springer, T.A.(2010) Blood 116: 5050-5059
- PubMed: 20679525
- DOI: https://doi.org/10.1182/blood-2010-04-281154
- Primary Citation of Related Structures:
3NID, 3NIF, 3NIG - PubMed Abstract:
The platelet integrin α(IIb)β(3) is essential for hemostasis and thrombosis through its binding of adhesive plasma proteins. We have determined crystal structures of the α(IIb)β(3) headpiece in the absence of ligand and after soaking in RUC-1, a novel small molecule antagonist. In the absence of ligand, the α(IIb)β(3) headpiece is in a closed conformation, distinct from the open conformation visualized in presence of Arg-Gly-Asp (RGD) antagonists. In contrast to RGD antagonists, RUC-1 binds only to the α(IIb) subunit. Molecular dynamics revealed nearly identical binding. Two species-specific residues, α(IIb) Y190 and α(IIb) D232, in the RUC-1 binding site were confirmed as important by mutagenesis. In sharp contrast to RGD-based antagonists, RUC-1 did not induce α(IIb)β(3) to adopt an open conformation, as determined by gel filtration and dynamic light scattering. These studies provide insights into the factors that regulate integrin headpiece opening, and demonstrate the molecular basis for a novel mechanism of integrin antagonism.
- Immune Disease Institute, Children's Hospital Boston, and Department of Pathology, Harvard Medical School, Boston, MA 02115, USA.
Organizational Affiliation: