The 2.2-A Structure of the HP0958 Protein from Helicobacter pylori Reveals a Kinked Anti-Parallel Coiled-Coil Hairpin Domain and a Highly Conserved Zn-Ribbon Domain
Caly, D.L., O'Toole, P.W., Moore, S.A.(2010) J Mol Biology 403: 405-419
- PubMed: 20826163
- DOI: https://doi.org/10.1016/j.jmb.2010.08.051
- Primary Citation of Related Structures:
3NA7 - PubMed Abstract:
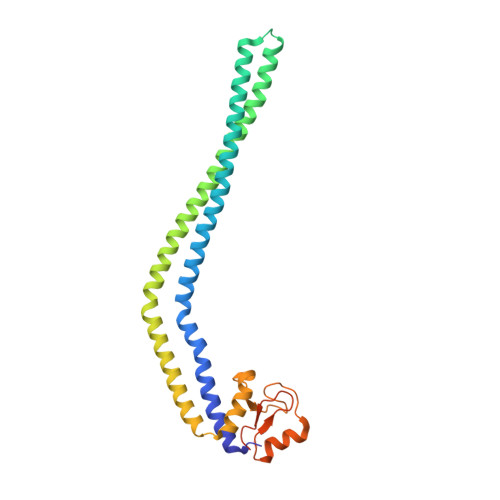
We have determined the 2.2-Å structure of the HP0958 protein from the human gastric pathogen Helicobacter pylori. HP0958 is essential for flagellum formation and motility. It functions as a chaperone for RpoN (σ(54)) and also controls the stability and translation of mRNA for the major flagellin subunit FlaA. The protein is composed of a highly elongated and kinked coiled-coil hairpin domain (residues 1-170), followed by a C(4) Zn-ribbon domain (residues 174-238). The Zn-ribbon domain is rich in aromatic and positively charged amino acid residues. Electrophoretic mobility shift assays identified residues in a positively charged region of the Zn-ribbon domain of HP0958 whose mutation alters the mobility of an HP0958-flaA mRNA complex. Mutation of surface residues in the coiled-coil domain did not result in an observable change in the mobility of the HP0958-flaA transcript complex. The data thus suggest the arrangement of HP0958 into distinct structural and functional domains.
- Department of Microbiology, University College Cork, Cork, Ireland.
Organizational Affiliation: