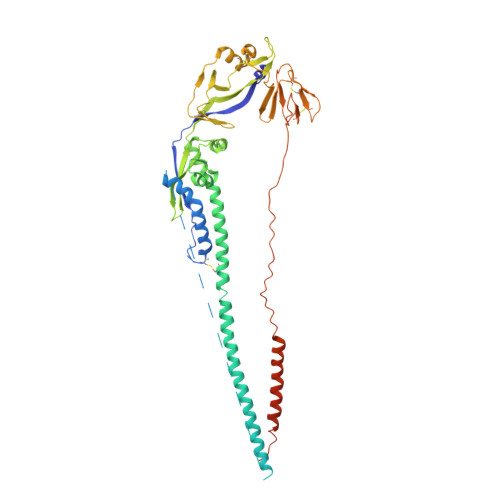
Structure of the Newcastle disease virus F protein in the post-fusion conformation.
Swanson, K., Wen, X., Leser, G.P., Paterson, R.G., Lamb, R.A., Jardetzky, T.S.(2010) Virology 402: 372-379
- PubMed: 20439109
- DOI: https://doi.org/10.1016/j.virol.2010.03.050
- Primary Citation of Related Structures:
3MAW - PubMed Abstract:
The paramyxovirus F protein is a class I viral membrane fusion protein which undergoes a significant refolding transition during virus entry. Previous studies of the Newcastle disease virus, human parainfluenza virus 3 and parainfluenza virus 5 F proteins revealed differences in the pre- and post-fusion structures. The NDV Queensland (Q) F structure lacked structural elements observed in the other two structures, which are key to the refolding and fusogenic activity of F. Here we present the NDV Australia-Victoria (AV) F protein post-fusion structure and provide EM evidence for its folding to a pre-fusion form. The NDV AV F structure contains heptad repeat elements missing in the previous NDV Q F structure, forming a post-fusion six-helix bundle (6HB) similar to the post-fusion hPIV3 F structure. Electrostatic and temperature factor analysis of the F structures points to regions of these proteins that may be functionally important in their membrane fusion activity.
- Howard Hughes Medical Institute, USA.
Organizational Affiliation: