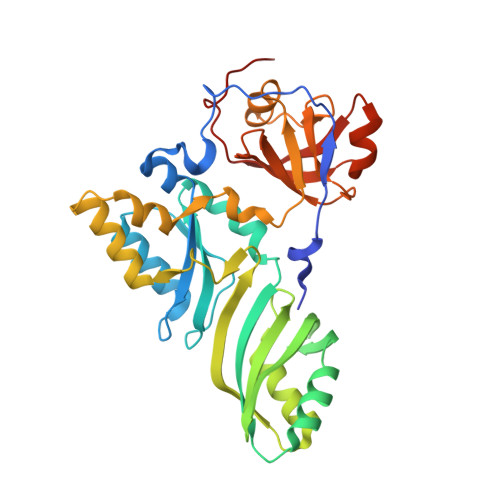
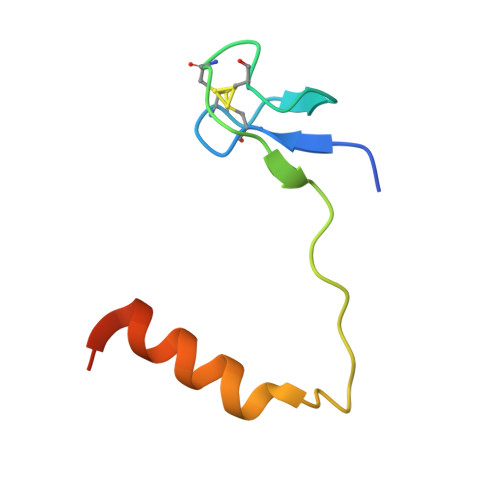
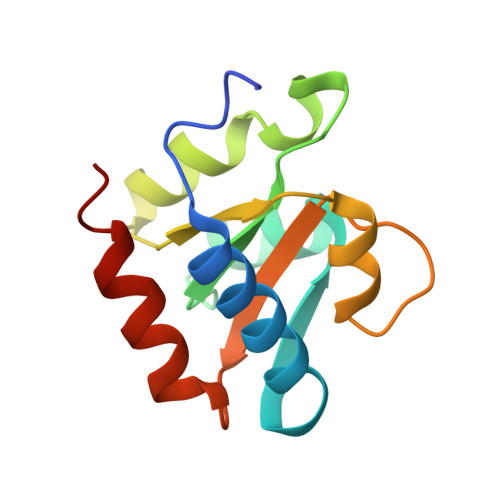
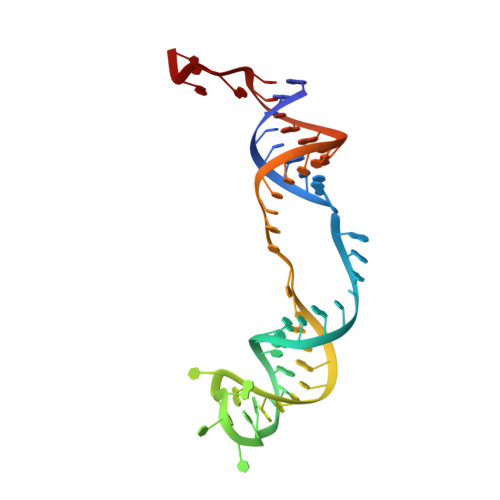
Glycosidic bond conformation preference plays a pivotal role in catalysis of RNA pseudouridylation: a combined simulation and structural study.
Zhou, J., Lv, C., Liang, B., Chen, M., Yang, W., Li, H.(2010) J Mol Biology 401: 690-695
- PubMed: 20615421
- DOI: https://doi.org/10.1016/j.jmb.2010.06.061
- Primary Citation of Related Structures:
3LWO, 3LWP - PubMed Abstract:
The most abundant chemical modification on RNA is isomerization of uridine (or pseudouridylation) catalyzed by pseudouridine synthases. The catalytic mechanism of this essential process remains largely speculative, partly due to lack of knowledge of the pre-reactive state that is important to the identification of reactive chemical moieties. In the present study, we showed, using orthogonal space random-walk free-energy simulation, that the pre-reactive states of uridine and its reactive derivative 5-fluorouridine, bound to a ribonucleoprotein particle pseudouridine synthase, strongly prefer the syn glycosidic bond conformation, while that of the nonreactive 5-bromouridine-containing substrate is largely populated in the anti conformation state. A high-resolution crystal structure of the 5-bromouridine-containing substrate bound to the ribonucleoprotein particle pseudouridine synthase and enzyme activity assay confirmed the anti nonreactive conformation and provided the molecular basis for its confinement. The observed preference for the syn pre-reactive state by the enzyme-bound uridine may help to distinguish among currently proposed mechanisms.
- Department of Chemistry and Biochemistry, Florida State University, Tallahassee, FL 32306, USA.
Organizational Affiliation: