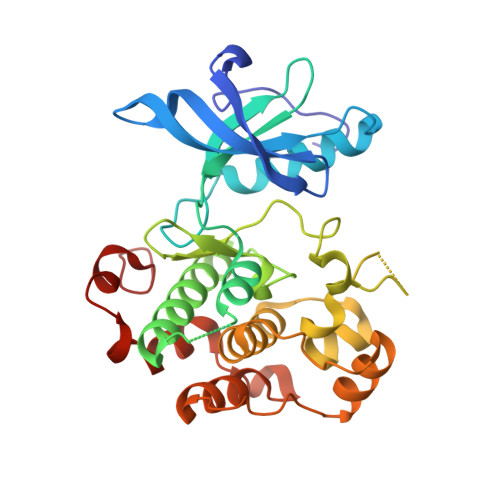
Allosteric IGF-1R Inhibitors.
Heinrich, T., Gradler, U., Bottcher, H., Blaukat, A., Shutes, A.(2010) ACS Med Chem Lett 1: 199-203
- PubMed: 24900194
- DOI: https://doi.org/10.1021/ml100044h
- Primary Citation of Related Structures:
3LW0 - PubMed Abstract:
Targeting allosteric protein sites is a promising approach to interfere selectively with cellular signaling cascades. We have discovered a novel class of allosteric insulin-like growth factor-I receptor (IGF-1R) inhibitors. 3-Cyano-1H-indole-7-carboxylic acid {1-[4-(5-cyano-1H-indol-3-yl)butyl]piperidin-4-yl}amide (10) was found with nanomolar biochemical, micromolar, cellular IGF-1R activity and no relevant interference with cellular insulin receptor signaling up to 30 μM. The allosteric binding site was characterized by X-ray crystallographic studies, and the structural information was used to explain the unique mode of action of this new class of inhibitors.
- Merck Serono, Frankfurter Strasse 250, 64293 Darmstadt, Germany.
Organizational Affiliation: