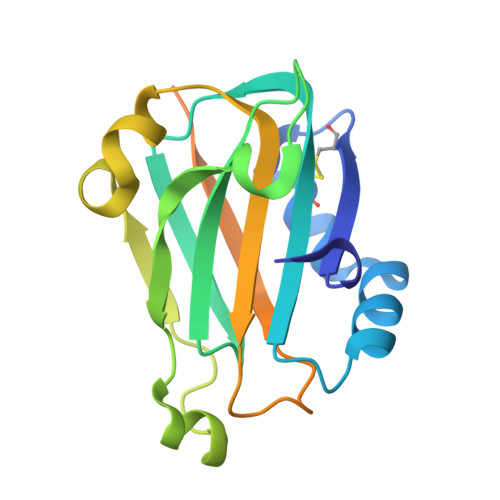
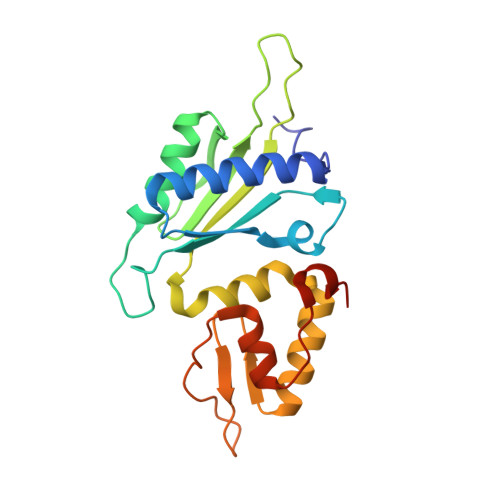
Oxidation state of the XRCC1 N-terminal domain regulates DNA polymerase beta binding affinity.
Cuneo, M.J., London, R.E.(2010) Proc Natl Acad Sci U S A 107: 6805-6810
- PubMed: 20351257
- DOI: https://doi.org/10.1073/pnas.0914077107
- Primary Citation of Related Structures:
3K75, 3K77, 3LQC - PubMed Abstract:
Formation of a complex between the XRCC1 N-terminal domain (NTD) and DNA polymerase beta (Pol beta) is central to base excision repair of damaged DNA. Two crystal forms of XRCC1-NTD complexed with Pol beta have been solved, revealing that the XRCC1-NTD is able to adopt a redox-dependent alternate fold, characterized by a disulfide bond, and substantial variations of secondary structure, folding topology, and electrostatic surface. Although most of these structural changes occur distal to the interface, the oxidized XRCC1-NTD forms additional interactions with Pol beta, enhancing affinity by an order of magnitude. Transient disulfide bond formation is increasingly recognized as an important molecular regulatory mechanism. The results presented here suggest a paradigm in DNA repair in which the redox state of a scaffolding protein plays an active role in organizing the repair complex.
- Laboratory of Structural Biology, National Institute of Environmental Health Sciences, Research Triangle Park, NC 27709, USA.
Organizational Affiliation: