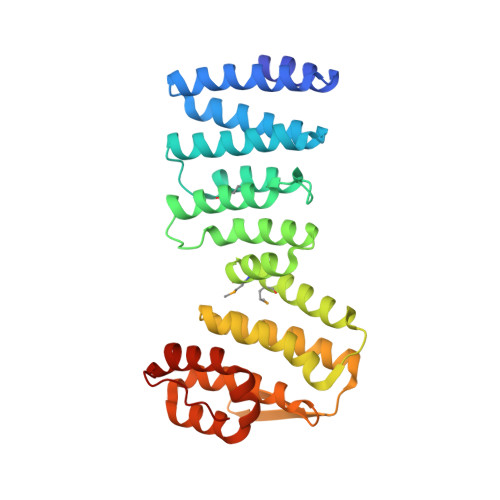
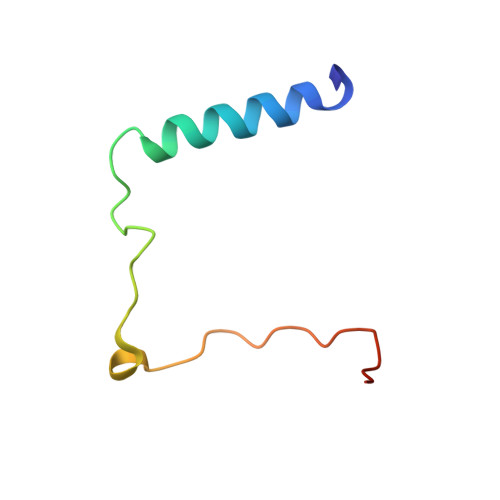
Structural characterization of the Get4/Get5 complex and its interaction with Get3.
Chartron, J.W., Suloway, C.J., Zaslaver, M., Clemons, W.M.(2010) Proc Natl Acad Sci U S A 107: 12127-12132
- PubMed: 20554915
- DOI: https://doi.org/10.1073/pnas.1006036107
- Primary Citation of Related Structures:
3LKU - PubMed Abstract:
The recently elucidated Get proteins are responsible for the targeted delivery of the majority of tail-anchored (TA) proteins to the endoplasmic reticulum. Get4 and Get5 have been identified in the early steps of the pathway mediating TA substrate delivery to the cytoplasmic targeting factor Get3. Here we report a crystal structure of Get4 and an N-terminal fragment of Get5 from Saccharomyces cerevisae. We show Get4 and Get5 (Get4/5) form an intimate complex that exists as a dimer (two copies of Get4/5) mediated by the C-terminus of Get5. We further demonstrate that Get3 specifically binds to a conserved surface on Get4 in a nucleotide dependent manner. This work provides further evidence for a model in which Get4/5 operates upstream of Get3 and mediates the specific delivery of a TA substrate.
- Division of Chemistry and Chemical Engineering, California Institute of Technology, 1200 East California Boulevard, Pasadena, CA 91125, USA.
Organizational Affiliation: