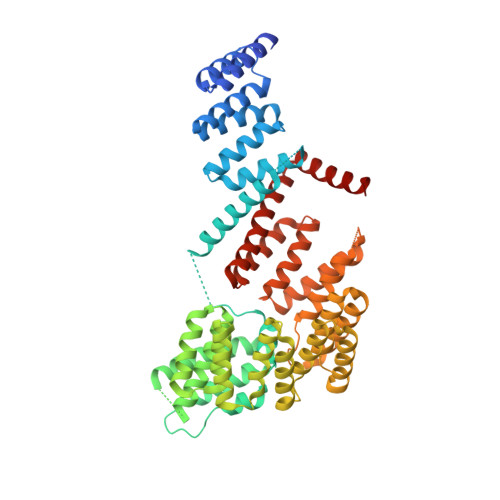
The structural plasticity of Tom71 for mitochondrial precursor translocations.
Li, J., Cui, W., Sha, B.(2010) Acta Crystallogr Sect F Struct Biol Cryst Commun 66: 985-989
- PubMed: 20823510
- DOI: https://doi.org/10.1107/S1744309110025522
- Primary Citation of Related Structures:
3LCA - PubMed Abstract:
Mitochondrial precursors are transported through the translocase of the outer membrane (TOM) complex. Tom70/Tom71 is a major surface receptor of the TOM complex for mitochondrial precursors and facilitates Hsp70/Hsp90-escorted precursor translocation into the mitochondrion. Previous structural studies of Tom71 have revealed that it contains an N-terminal and a C-terminal domain and that the two domains may remain in an open conformation when binding to Hsp70/Hsp90. In a newly obtained crystal form of a complex of Tom71 and the Hsp70 C-terminus, the N-terminal domain was found to have rotated about 12 degrees towards the C-terminal domain compared with the previous determined crystal structure of Tom71 in the open conformation. This newly solved structure is defined as the ;intermediate conformation'. The domain rearrangements in Tom71 significantly change the surface hydrophobicity and the volume of the precursor-binding pocket. This work suggests that Tom70/Tom71-family members may exhibit structural plasticity from the intermediate conformation to the fully open conformation when complexed with Hsp70/Hsp90. This structural plasticity enables the precursor receptors to accommodate different precursor substrates for mitochondrial translocation.
- Department of Cell Biology, University of Alabama at Birmingham, Birmingham, AL 35294, USA.
Organizational Affiliation: