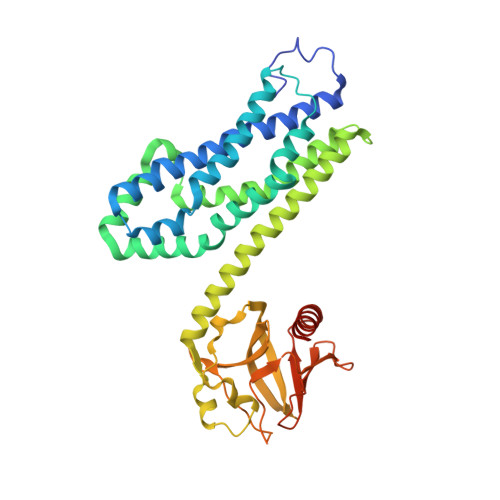
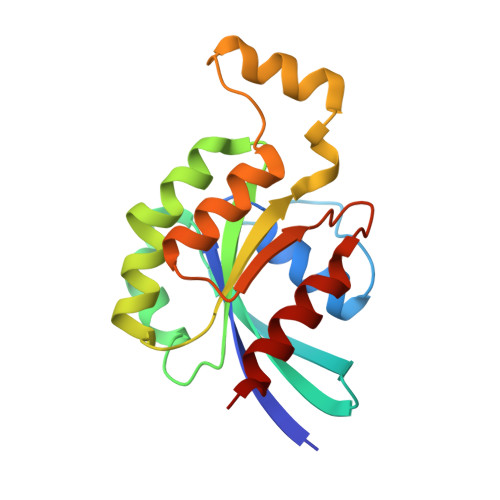
Activated RhoA binds to the pleckstrin homology (PH) domain of PDZ-RhoGEF, a potential site for autoregulation.
Chen, Z., Medina, F., Liu, M.Y., Thomas, C., Sprang, S.R., Sternweis, P.C.(2010) J Biological Chem 285: 21070-21081
- PubMed: 20430886
- DOI: https://doi.org/10.1074/jbc.M110.122549
- Primary Citation of Related Structures:
3KZ1 - PubMed Abstract:
Guanine nucleotide exchange factors (GEFs) catalyze exchange of GDP for GTP by stabilizing the nucleotide-free state of the small GTPases through their Dbl homology/pleckstrin homology (DH.PH) domains. Unconventionally, PDZ-RhoGEF (PRG), a member of the RGS-RhoGEFs, binds tightly to both nucleotide-free and activated RhoA (RhoA.GTP). We have characterized the interaction between PRG and activated RhoA and determined the structure of the PRG-DH.PH-RhoA.GTPgammaS (guanosine 5'-O-[gamma-thio]triphosphate) complex. The interface bears striking similarity to a GTPase-effector interface and involves the switch regions in RhoA and a hydrophobic patch in PRG-PH that is conserved among all Lbc RhoGEFs. The two surfaces that bind activated and nucleotide-free RhoA on PRG-DH.PH do not overlap, and a ternary complex of PRG-DH.PH bound to both forms of RhoA can be isolated by size-exclusion chromatography. This novel interaction between activated RhoA and PH could play a key role in regulation of RhoGEF activity in vivo.
- Department of Pharmacology, The University of Texas Southwestern Medical Center, Dallas, TX 75390, USA. Zhe.Chen@utsouthwestern.edu
Organizational Affiliation: